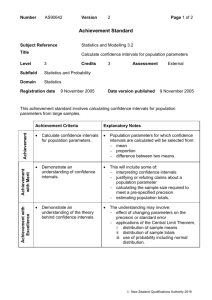
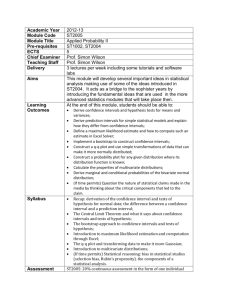
Galaxy-History_2-Single-Copy Stable Sequence Determination
advertisement

We used Galaxy to retrieve FASTA sequences of single-copy (sc-) stable intervals. Subsequently, these sequences were used in mpiBLAST for determining multi-copy stable intervals. This is a Galaxy history for chromosome 18 as an example of the workflow used for retrieving sc-stable sequences for all autosomes. Galaxy File: (insert) Dataset 1 2 Dataset Description Uploaded stable regions Uploaded repeat masker file from UCSC 3 Uploaded segmental duplication track from UCSC 4 Filtered data by chromosome of interest Subtracted repeats from all stable regions --> result: list of single copy (sc) stable intervals Determined sc-stable intervals having segmental duplicons by performing an intersection Determined if there are any overlapping intervals that have segmental duplicons Subtracted clustered sc-stable intervals with segmental duplications - to return NON-overlapping intervals that have segmental duplications Formatted the list of sc-stable intervals having segmental duplications Computed a subtraction: column3 - column2 to determine the length of the sc-stable intervals Filtered the results by keeping intervals having length of >500bp Extract FASTA sequence files for each sc-stable interval (hg16 assembly) Converted genomic coordinates of sc-stable intervals (>500bp) to hg18 assembly Extracted FASTA sequences for each sc-stable interval (>500bp) hg18 Re-filtered the data by length of sc-stable interval: by >100bp Converted all sc-stable intervals to hg18 assembly (>100bp) This determined how many and which sc-stable intervals (>100bp and <500bp) were not used in mpiBLAST search (as only >500bp intervals were used) 5 6 7 8 9 10 11 12 13-14 15 16 17-18 19 Galaxy Tool Upload File UCSC Main Table Browser: rmsk UCSC Main Table Browser: genomicSuperDups Filter on data 3 Subtract on data 1 and data 2 Intersect on data 4 and data 5 Cluster on data 6 Subtract on data 6 and data 7 Cut on data 8 Compute on data 9 Filter on data 10 Extract Genomic DNA on data 11 Convert genome coordinates on data 11 Extract Genomic DNA on data 14 Filter on data 10 Convert genome coordinates on data 16 Join on data 14 and data 18

![The Average rate of change of a function over an interval [a,b]](http://s3.studylib.net/store/data/005847252_1-7192c992341161b16cb22365719c0b30-300x300.png)


