Appendix 3_mar2015
advertisement

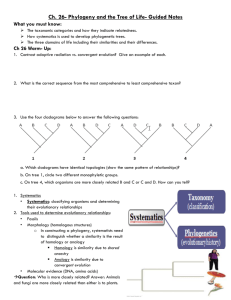
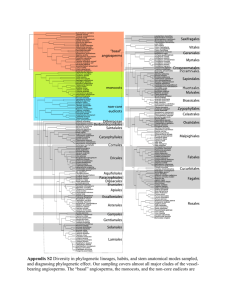
APPENDIX 3 SIMULATION EXPERIMENT DEMONSTRATING THE EQUIVALENCE BETWEEN THE CENSORED APPROACH (O’MEARA ET AL. 2006) AND THE RATE METHOD USING A DESIGN MATRIX FOR GROUP MEANS (THOMAS ET AL. 2006, 2009) Maximum-likelihood methods for comparing evolutionary rates across groups defined by a discrete, binary trait across a phylogeny were simultaneously developed by O’Meara et al. (2006) and Thomas et al. (2006). Both studies proposed slightly different models for rate comparison. O’Meara et al. (2006) proposed two different approaches. In the “non-censored” approach, each branch on the tree is assigned a rate parameter based on a single phylogenetic mean for the whole tree (e.g. the expected value E(X), corresponding to the phenotypic value at the root). In contrast, the “censored” approach works by eliminating the branches not included in each of the defined groups and calculating a rate estimate based on this reduced tree, also calculating a separate phylogenetic mean for each group (O’Meara et al. 2006). This approach is more flexible in terms of the assumptions made regarding the exact point where a rate shift occurred, but is restricted to monophyletic subtrees. Simultaneously, Thomas et al. (2006) proposed a similar method, where rate comparison between groups is performed using a design matrix to calculate the expected values (phylogenetic means) for each group (Thomas et al. 2006, 2009). While the model of Thomas et al. 2006 allows for any type of distribution of groups across the tree (e.g. there is no restriction to monophyly), it coincides with the censored approach in using different phylogenetic means for each of the examined groups. As such, both models are mathematically expected to have equal likelihoods and provide the same relative rate estimates in cases when they are comparable, e.g. when at least one of the examined groups is monophyletic. However, the identity of these two methods has never been formally tested. 1 We used a simulation experiment to confirm whether this was in fact the case. Simulations were conducted on a random phylogeny with 64 species, divided in two groups originating in a random node relatively deep in the tree, where at least one of the groups was monophyletic. We then used the transformPhylo.sim function of R-package motmot (Thomas and Freckleton 2012) to simulate a continuous phenotypic trait evolving under a BM with varying degrees of mean and rate differentiation between groups. The mean for the first group was always set to â1 = 0 and the rate of the first group was always set to σ21 = 1. The mean for the second group was set to either 0.5, 1, 2, 3, 4, 5, or 6 standard deviations larger than â1, whereas the rate of the second group (σ22 = θσ21) was set to: 1.1, 1.5, 2, 3, 4, 5, or 6 times larger than σ21. For each of the above combinations, 1000 phenoytpic datasets were simulated. We then used the censored approach, as implemented in R-package RBrownie (Stack et al. 2011), and the function ML.RatePhylo of motmot to fit the two evolutionary models for rate comparison mentioned above. Finally, we compared the obtained relative rate estimates (θcen and θmotmot), as well as the corresponding model log-likelihoods (loglikcen and loglikmotmot, by examining the correlation across 1000 datasets and the mean ratio of the values obtained from each implementation. The results obtained clearly demonstrate a full correspondence between the two methods when comparing evolutionary rates for continuous phenotypic traits between monophyletic groups on a phylogeny. Across 1000 simulated datasets, and across varying conditions of differentiation in phylogenetic means and rates between groups, the correlation and mean ratio of both relative rate estimates and log-likelihoods obtained by the two methods approached unit value (Table A3, Fig. A3), showing that the two implementations are fully coincident. 2 Relative rate (θ) Relative rate (θ) Relative rate (θ) Relative rate (θ) Table A3: Correlations and mean ratios of relative rate estimates and likelihoods obtained from fitting a two-rate model using the censored approach (O’Meara et al. 2006) or the rate test using a group design matrix for phylogenetic means (Thomas et al. 2006) across different simulation conditions for 1000 datasets simulated under a two-means, two-rates BM process. Relative rate estimates Difference in means (in St.Devs) Difference in means (in St.Devs) Correlation Ratio 0.5 1 2 3 4 5 6 0.5 1 2 3 4 5 6 1.1 1.1 1.020 1.020 1.020 1.020 1.020 1.020 1.021 1.000 1.000 1.000 1.000 1.000 1.000 1.000 1.5 1.5 1.024 1.024 1.024 1.024 1.024 1.024 1.024 1.000 1.000 1.000 1.000 1.000 1.000 1.000 2 2 1.027 1.028 1.027 1.028 1.027 1.027 1.028 1.000 1.000 1.000 1.000 1.000 1.000 1.000 3 3 1.032 1.032 1.032 1.032 1.032 1.032 1.032 1.000 1.000 1.000 0.999 1.000 0.999 1.000 4 4 1.035 1.035 1.035 1.035 1.035 1.035 1.035 0.999 1.000 1.000 1.000 1.000 1.000 1.000 5 5 1.037 1.037 1.037 1.037 1.037 1.037 1.037 1.000 0.999 1.000 1.000 1.000 1.000 1.000 6 6 1.039 1.038 1.038 1.039 1.038 1.038 1.038 1.000 1.000 1.000 1.000 1.000 1.000 0.999 Log-likelihoods Difference in means (in St.Devs) Difference in means (in St.Devs) Correlation Ratio 0.5 1 2 3 4 5 6 0.5 1 2 3 4 5 6 1.1 1.1 1.110 1.141 1.144 1.121 1.184 1.115 1.123 0.997 0.997 0.997 0.997 0.997 0.997 0.997 1.5 1.5 1.089 1.033 1.060 1.206 1.146 1.080 1.113 0.998 0.998 0.998 0.998 0.998 0.998 0.998 2 2 1.066 1.063 1.164 1.066 1.065 1.065 1.066 0.998 0.998 0.998 0.998 0.998 0.998 0.998 3 3 1.048 1.059 1.060 1.021 1.058 1.057 1.055 0.999 0.999 0.999 0.999 0.999 0.999 0.999 4 4 1.050 1.055 1.046 1.059 1.046 1.043 1.053 0.999 0.999 0.999 0.999 0.999 0.999 0.999 5 5 1.053 1.048 1.003 1.034 1.049 1.046 1.043 1.000 1.000 1.000 1.000 1.000 1.000 1.000 6 6 1.048 0.994 1.043 1.038 1.044 1.010 1.060 1.000 1.000 1.000 1.000 1.000 1.000 1.000 3 Figure A3: Comparison of relative rate estimates and log-likelihoods obtained from the censored test (θcen and loglikcen) and from ML.RatePhylo (θmotmot and loglikmotmot). Results are shown for the case of â2 / â1 = 4*stdev and θ=4, but were similar for all the simulation conditions examined (Table A3). References Stack, C.J, Harmon, L.J. & O’Meara, B. 2011. RBrownie: an R package for testing hypotheses about rates of evolutionary change. Methods in Ecology and Evolution 2: 660-662. 4