Section 16
advertisement

Logistic Regression in R
In this section of the notes we examine logistic regression in R. There are several
functions that I wrote for plotting diagnostics similar to what SAS does, although the
inspiration for them came from work Prof. Malone and I did for OLS as part of his senior
project.
Example 1: Oral Contraceptive Use and Myocardial Infarctions
Set up a text file with the data in columns with variable names at the top. The case and control
counts are in separate columns. The risk factor OC use and stratification variable Age follow.
> OCMI.data = read.table(file.choose(),header=T)
# read in text file
> OCMI.data
MI NoMI Age OCuse
1
4
62
1
Yes
2
2 224
1
No
3
9
33
2
Yes
4 12 390
2
No
5
4
26
3
Yes
6 33 330
3
No
7
6
9
4
Yes
8 65 362
4
No
9
6
5
5
Yes
10 93 301
5
No
> attach(OCMI.data)
> OC.glm <- glm(cbind(MI,NoMI)~Age+OCuse,family=binomial)
# fit model
> summary(OC.glm)
Call:
glm(formula = cbind(MI, NoMI) ~ Age + OCuse, family = binomial)
Deviance Residuals:
[1]
0.456248 -0.520517
[9] -0.045061
0.008822
1.377693
-0.886710
-1.685521
Coefficients:
Estimate Std. Error z value
(Intercept) -4.3698
0.4347 -10.054
Age2
1.1384
0.4768
2.388
Age3
1.9344
0.4582
4.221
Age4
2.6481
0.4496
5.889
Age5
3.1943
0.4474
7.140
OCuseYes
1.3852
0.2505
5.530
--Signif. codes: 0 `***' 0.001 `**' 0.01
0.714695
Pr(>|z|)
< 2e-16
0.0170
2.43e-05
3.88e-09
9.36e-13
3.19e-08
-0.130922
0.033643
***
*
***
***
***
***
`*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 158.0085
Residual deviance:
6.5355
AIC: 58.825
on 9
on 4
degrees of freedom
degrees of freedom
54
Number of Fisher Scoring iterations: 3
Find OR associated with oral contraceptive use ADJUSTED for age.
Recall: CMH procedure gave 3.97.
> exp(1.3852)
[1] 3.995625
Find a 95% CI for OR associated with OC use.
> exp(1.3852-1.96*.2505)
[1] 2.445428
> exp(1.3852+1.96*.2505)
[1] 6.528518
Interpreting the age effect in terms of OR’s ADJUSTING for OC use.
Note: The reference group is Age = 1 which was women 25 – 29 years of age.
> OC.glm$coefficients
(Intercept)
Age2
-4.369850
1.138363
Age3
1.934401
Age4
2.648059
Age5
3.194292
OCuseYes
1.385176
> Age.coefs <- OC.glm$coefficients[2:5]
> exp(Age.coefs)
Age2
Age3
Age4
Age5
3.121653 6.919896 14.126585 24.392906
Find 95% CI for age = 5 group.
> exp(3.1943-1.96*.4474)
[1] 10.14921
> exp(3.1943+1.96*.4474)
[1] 58.62751
Example 2: Coffee Drinking and Myocardial Infarctions
CoffeeMI.data = read.table(file.choose(),header=T)
> CoffeeMI.data
Smoking Coffee MI NoMI
1
Never
> 5 7
31
2
Never
< 5 55 269
3
Former
> 5 7
18
4
Former
< 5 20 112
5
1-14 Cigs
> 5 7
24
6
1-14 Cigs
< 5 33 114
7 15-25 Cigs
> 5 40
45
8 15-25 Cigs
< 5 88 172
9 25-34 Cigs
> 5 34
24
10 25-34 Cigs
< 5 50
55
11 35-44 Cigs
> 5 27
24
12 35-44 Cigs
< 5 55
58
13
45+ Cigs
> 5 30
17
14
45+ Cigs
< 5 34
17
> attach(CoffeeMI.data)
> Coffee.glm = glm(cbind(MI,NoMI)~Smoking+Coffee,family=binomial)
55
> summary(Coffee.glm)
Call:
glm(formula = cbind(MI, NoMI) ~ Smoking + Coffee, family = binomial)
Deviance Residuals:
Min
1Q
Median
-0.7650 -0.4510 -0.0232
3Q
0.2999
Max
0.7917
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept)
-1.2981
0.1819 -7.136 9.60e-13 ***
Smoking15-25 Cigs
0.6892
0.2119
3.253 0.00114 **
Smoking25-34 Cigs
1.2462
0.2398
5.197 2.02e-07 ***
Smoking35-44 Cigs
1.1988
0.2389
5.017 5.24e-07 ***
Smoking45+ Cigs
1.7811
0.2808
6.342 2.27e-10 ***
SmokingFormer
-0.3291
0.2778 -1.185 0.23616
SmokingNever
-0.3153
0.2279 -1.384 0.16646
Coffee> 5
0.3200
0.1377
2.324 0.02012 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 173.7899
Residual deviance:
3.7622
AIC: 84.311
on 13
on 6
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 3
OR for drinking 5 or more cups of coffee per day.
Note: CMH procedure gave OR = 1.375
> exp(.3200)
[1] 1.377128
95% CI for OR associated with heavy coffee drinking
> exp(.3200 - 1.96*.1377)
[1] 1.051385
> exp(.3200 + 1.96*.1377)
[1] 1.803794
Reordering a Factor
To examine the effect of smoking we might want to “reorder” the levels of smoking
status so that individuals who have never smoked are used as the reference group. To do
this in R you must do the following:
Smoking = factor(Smoking,levels=c("Never","Former","1-14 Cigs","15-25
Cigs","25-34 Cigs","35-44 Cigs","45+ Cigs"))
The first level specified in the levels subcommand will be used as the reference group,
“Never” in this case. Refitting the model with the reordered smoking status factor gives
the following:
56
> Coffee.glm2 <-glm(cbind(MI,NoMI)~Smoking+Coffee,family=binomial)
> summary(Coffee.glm2)
Call:
glm(formula = cbind(MI, NoMI) ~ Smoking + Coffee, family = binomial)
Deviance Residuals:
Min
1Q
Median
3Q
Max
-0.7650 -0.4510 -0.0232
0.2999
0.7917
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept)
-1.61344
0.14068 -11.469 < 2e-16 ***
SmokingFormer
-0.01376
0.25376 -0.054
0.9568
Smoking1-14 Cigs
0.31533
0.22789
1.384
0.1665
Smoking15-25 Cigs 1.00451
0.17976
5.588 2.30e-08 ***
Smoking25-34 Cigs 1.56150
0.21254
7.347 2.03e-13 ***
Smoking35-44 Cigs 1.51417
0.21132
7.165 7.77e-13 ***
Smoking45+ Cigs
2.09646
0.25855
8.108 5.13e-16 ***
Coffee> 5
0.31995
0.13766
2.324
0.0201 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 173.7899
Residual deviance:
3.7622
AIC: 84.311
on 13
on 6
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 3
Notice that “SmokingNever” is now absent from the output so we know it is being used
as the reference group. The OR’s associated with the various levels of smoking are
computed below.
> Smoke.coefs = Coffee.glm$coefficients[2:7]
> exp(Smoke.coefs)
SmokingFormer Smoking1-14 Cigs Smoking15-25 Cigs Smoking25-34 Cigs
0.986338
1.370715
2.730561
4.765984
Smoking35-44 Cigs
Smoking45+ Cigs
4.545632
8.137279
Confidence intervals for each could be computed in the standard way.
57
Some Details for Categorical Predictors with More Than Two Levels
Consider the coffee drinking/MI study above. The stratification variable smoking has
seven levels. Thus it requires six dummy variables to define it. The level that is not
defined using a dichotomous dummy variable serves as the reference group. The table
below shows how the value of the dummy variables:
Level
D2
D3
D4
D5
D6
D7
Never
0
0
0
0
0
0
1
0
0
0
0
0
0
1
0
0
0
0
0
0
1
0
0
0
0
0
0
1
0
0
0
0
0
0
1
0
0
0
0
0
0
1
(Reference
Group)
Former
1 – 14 Cigs
15 – 24 Cigs
25 – 34 Cigs
35 – 44 Cigs
45+ Cigs
Example: Coffee Drinking and Myocardial Infarctions
CoffeeMI.data = read.table(file.choose(),header=T)
> CoffeeMI.data
Smoking Coffee MI NoMI
1
Never
> 5 7
31
2
Never
< 5 55 269
3
Former
> 5 7
18
4
Former
< 5 20 112
5
1-14 Cigs
> 5 7
24
6
1-14 Cigs
< 5 33 114
7 15-25 Cigs
> 5 40
45
8 15-25 Cigs
< 5 88 172
9 25-34 Cigs
> 5 34
24
10 25-34 Cigs
< 5 50
55
11 35-44 Cigs
> 5 27
24
12 35-44 Cigs
< 5 55
58
13
45+ Cigs
> 5 30
17
14
45+ Cigs
< 5 34
17
The Logistic Model
( x)
Coffee D D D D D D
~
ln
o
1
2 2
3 3
4 4
5 5
6 6
7 7
1 ( x)
~
where Coffee is a dichotomous predictor equal to 1 if they drink 5 or more cups of coffee
per day.
Comparing the log-odds of a heavy coffee drinker who who smokes 15-25 cigarettes day
to a heavy coffee drinker who has never smoked we have.
58
1 ( x)
~
ln
o
1
4
1 1 ( x)
~
2 ( x)
~
ln
o
1
1 2 ( x)
~
Taking the difference gives,
1 ( x)
~
1 1 ( x)
~
ln
4
2 ( x~ )
1 2 ( x)
~
thus
e 4 the odds ratio associated with smoking 15-24 cigarettes per day when compared to
individuals who have never smoked amongst heavy coffee drinkers. Because 1 is not
involved in the odds ratio the result is the same for non-heavy coffee drinkers as well!
You can also consider combinations of factors, e.g. if we compared heavy coffee drinkers
who smoked 15-24 cigarettes to a non-heavy coffee drinkers who have never smoked the
associated OR would be given by e1 4 .
Using our fitted model the OR’s ratios discussed above would be.
> summary(Coffee.glm)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept)
-1.61344
0.14068 -11.469 < 2e-16 ***
SmokingFormer
-0.01376
0.25376 -0.054
0.9568
Smoking1-14 Cigs
0.31533
0.22789
1.384
0.1665
Smoking15-25 Cigs 1.00451
0.17976
5.588 2.30e-08 ***
Smoking25-34 Cigs 1.56150
0.21254
7.347 2.03e-13 ***
Smoking35-44 Cigs 1.51417
0.21132
7.165 7.77e-13 ***
Smoking45+ Cigs
2.09646
0.25855
8.108 5.13e-16 ***
Coffee> 5
0.31995
0.13766
2.324
0.0201 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
OR for 15-24 cigarette smokers vs. never smokers (regardless of coffee drinking status)
> exp(1.00451)
[1] 2.730569
59
OR for 15-24 cigarette smokers who are also heavy coffee drinkers vs. non-smokers who
are not heavy coffee drinkers
> exp(.31995 + 1.00451)
[1] 3.760154
Similar calculations could be done for other combinations of coffee and cigarette use.
Example 3: Risk Factors for Low Birth Weight
Response
Y = low birth weight, i.e. birth weight < 2500 grams (1 = yes, 0 = no)
Set of potential predictors
X1
X2
X3
X4
X5
X6
X7
=
=
=
=
=
=
=
previous history of premature labor (1 = yes, 0 = no)
hypertension during pregnancy (1 = yes, 0 = no)
smoker (1 = yes, 0 = no)
uterine irritability (1 = yes, 0 = no)
minority (1 = yes, 0 = no)
mother’s age in years
mother’s weight at last menstrual cycle
Analysis in R
> Lowbirth = read.table(file.choose(),header=T)
> Lowbirth[1:5,]
# print first 5 rows of the data set
Low Prev Hyper Smoke Uterine Minority Age Lwt race bwt
1
0
0
0
0
1
1 19 182
2 2523
2
0
0
0
0
0
1 33 155
3 2551
3
0
0
0
1
0
0 20 105
1 2557
4
0
0
0
1
1
0 21 108
1 2594
5
0
0
0
1
1
0 18 107
1 2600
Make sure categorical variables are interpreted as factors by using the factor command
>
>
>
>
>
>
Low = factor(Low)
Prev = factor(Prev)
Hyper = factor(Hyper)
Smoke = factor(Smoke)
Uterine = factor(Uterine)
Minority = factor(Minority)
Note: This is not really necessary for dichotomous variables that are coded (0,1).
Fit a preliminary model using all available covariates
> low.glm = glm(Low~Prev+Hyper+Smoke+Uterine+Minority+Age+Lwt,family=binomial)
> summary(low.glm)
Call:
glm(formula = Low ~ Prev + Hyper + Smoke + Uterine + Minority +
Age + Lwt, family = binomial)
Deviance Residuals:
Min
1Q
Median
-1.6010 -0.8149 -0.5128
3Q
1.0188
Max
2.1977
60
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.378479
1.170627
0.323 0.74646
Prev1
1.196011
0.461534
2.591 0.00956 **
Hyper1
1.452236
0.652085
2.227 0.02594 *
Smoke1
0.959406
0.405302
2.367 0.01793 *
Uterine1
0.647498
0.466468
1.388 0.16511
Minority1
0.990929
0.404969
2.447 0.01441 *
Age
-0.043221
0.037493 -1.153 0.24900
Lwt
-0.012047
0.006422 -1.876 0.06066 .
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Null deviance: 232.40
Residual deviance: 196.71
AIC: 212.71
on 185
on 178
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 3
It appears that both uterine irritability and mother’s age are not significant. We can fit
the reduced model eliminating both terms and test whether the model is significantly
degraded by using the general chi-square test (see the SAS example).
> low.reduced = glm(Low~Prev+Hyper+Smoke+Minority+Lwt,family=binomial)
> summary(low.reduced)
Call:
glm(formula = Low ~ Prev + Hyper + Smoke + Minority + Lwt, family =
binomial)
Deviance Residuals:
Min
1Q
Median
-1.7277 -0.8219 -0.5368
3Q
0.9867
Max
2.1517
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.261274
0.885803 -0.295 0.76803
Prev1
1.181940
0.444254
2.661 0.00780 **
Hyper1
1.397219
0.656271
2.129 0.03325 *
Smoke1
0.981849
0.398300
2.465 0.01370 *
Minority1
1.044804
0.394956
2.645 0.00816 **
Lwt
-0.014127
0.006387 -2.212 0.02697 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 232.40
Residual deviance: 200.32
AIC: 212.32
on 185
on 180
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 3
61
( x)
~
X X X X X
H o : ln
o
1 1
2
2
3
3
5
5
7
7
1 ( x)
~
( x)
~
X X X X X X X
H 1 : ln
o
1 1
2
2
3
3
4
4
5
5
6
6
7
7
1 ( x)
~
* Recall: ( x) P( Low 1 | X )
~
~
DH o 200.32
df = 180
Residual Deviance Alternative Hypothesis Model: DH1 196.71
df = 178
Residual Deviance Null Hypothesis Model:
General Chi-Square Test
2 DH 0 DH1 200.32 196.71 3.607
p value P( 2 3.607) .1647
Fail to reject the null, the reduced model is adequate.
2
Interpretation of Model Parameters
OR’s Associated with Categorical Predictors
> low.reduced
Call: glm(formula = Low ~ Prev + Hyper + Smoke + Minority + Lwt,
family = binomial)
Coefficients:
(Intercept)
Prev1
-0.26127 1.18194
Hyper1
1.39722
Smoke1
0.98185
Degrees of Freedom: 185 Total (i.e. Null);
Null Deviance:
232.4
Residual Deviance: 200.3
AIC: 212.3
Minority1
1.04480
Lwt
-0.01413
180 Residual
Estimated OR’s
> exp(low.reduced$coefficients[2:5])
Prev1
Hyper1
Smoke1 Minority1
3.260693 4.043938 2.669388 2.842841
95% CI for OR Associated with History of Premature Labor
> exp(1.182 - 1.96*.444)
[1] 1.365827
> exp(1.182 + 1.96*.444)
[1] 7.78532
Holding everything else constant we estimate that the odds of having an infant with low birth
weight are between 1.366 and 7.785 times larger for mothers with a history of premature labor.
62
95% CI for OR Associated with Hypertension
> exp(1.397 - 1.96*.6563)
[1] 1.117006
> exp(1.397 + 1.96*.6563)
[1] 14.63401
Holding everything else constant we estimate that the odds of having an infant with low birth
weight are between 1.117 and 14.63 times larger for mothers with hypertension during
pregnancy.
95% CI for OR Associated with Smoking
> exp(.981849 - 1.96*.3983)
[1] 1.222846
> exp(.981849 + 1.96*.3983)
[1] 5.827086
Holding everything else constant we estimate that the odds of having an infant with low birth
weight are between 1.223 and 5.827 times larger for mothers who smoked during pregnancy.
95% CI for OR Associated with Minority Status
> exp(1.0448 - 1.96*.3950)
[1] 1.310751
> exp(1.0448 + 1.96*.3950)
[1] 6.16569
Holding everything else constant we estimate that the odds of having an infant with low birth
weight are between 1.311 and 6.166 times larger for non-white mothers.
OR Associated with Mother’s Weight at Last Menstrual Cycle
Because this is a continuous predictor with values over 100 we should use an increment
larger than one when considering the effect of mother’s weight on birth weight. Here we
will use an increment of c = 10 lbs., although certainly there are other possibilities.
> exp(-10*.014127)
[1] 0.8682549
i.e. 13.2% decrease in the OR for each additional 10 lbs. in premenstrual weight.
A 95% CI for this OR is:
> exp(10*(-.014127) - 1.96*10*.006387)
[1] 0.7660903
> exp(10*(-.014127) + 1.96*10*.006387)
[1] 0.9840439
Create a sequence of weights from smallest observed weight to the largest observed weight by ½
pound increments.
> x = seq(min(Lwt),max(Lwt),.5)
63
Here I have set the other covariates as follows: previous history (1 = yes), hypertension
(0 = no), smoking status (1 = yes), and minority (0 = no).
> fit = predict(low.reduced,data.frame(Prev=factor(rep(1,length(x))),
Hyper=factor(rep(0,length(x))),Smoke=factor(rep(1,length(x))),Minority=
factor(rep(0,length(x))),Lwt=x),type="response")
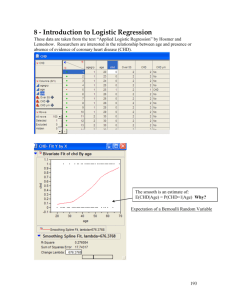
plot(x,fit,xlab=”Mother’s Weight”,ylab=”P(Low|Prev=1,Smoke=1,Lwt)”)
This is a plot of the effect of
premenstrual weight for smoking
mothers with a history of premature
labor. Using the predict command
above similar plots could be
constructed by examining other
combinations of the categorical
predictors.
64
Case Diagnostics (Delta Deviance and Cook’s Distance)
As in the case of ordinary least squares (OLS) regression we need to be wary of cases
that may have unduly high influence on our results and those that are poorly fit. The
most common influence measure is Cook’s Distance and a good measure of poorly fit
cases is the Delta Deviance.
Essentially Cook’s Distance ( ˆ( i ) or 𝐷𝑖 ) measures the changes in the estimated
parameters when the ith observation is deleted. This change is measured for each of the
observations and can be plotted versus ˆ( x) or observation number to aid in the
~
identification of high influence cases. Several cut-offs have been proposed for Cook’s
Distance, the most common being to classify an observation as having large influence if
ˆ( i ) 1 or, in case of large sample size n, ˆ( i ) 4 / n .
Cook’s Distance
( i )
2
1 eˆ i
k 1 hi
where eˆ χ i
hi
1 hi
y i yˆ i
is the Pearson’s residual defined above.
n i θˆ (~
x i )(1 θˆ (~
x i ))
Delta deviance measures the change in the deviance (D) when the ith case is deleted.
Values around 4 or larger are considered to cases that are poorly fit.
These cases correspond to cases to individuals where yi 1 but ˆ( x) is small, or cases
~
where yi 0 but ˆ( x) is large.
~
In cases of both high influence and poor fit it is good to look at the covariate values for
these individuals and we can begin to address the role they play in the analysis. In many
cases there will be several individuals with the same covariate pattern, especially if most
or all of the predictors are categorical in nature.
> Diagplot.glm(low.reduced)
65
> Diagplot.log(low.reduced)
Cases 11 and 13 have the highest Cook’s distances although they are not that large. It
should be noted also that they are also somewhat poorly fit. Cases 129, 144, 152, and
180 appear to be poorly fit. The information on all of these cases is shown below.
> Lowbirth[c(11,13,129,144,152,180),]
Low Prev Hyper Smoke Uterine Minority Age Lwt race bwt
11
0
0
1
0
0
1 19 95
3 2722
13
0
0
1
0
0
1 22 95
3 2750
129
1
0
0
0
1
0 29 130
1 1021
144
1
0
0
0
1
1 21 200
2 1928
152
1
0
0
0
0
0 24 138
1 2100
180
1
0
0
1
0
0 26 190
1 2466
66
Case 152 had a low birth weight infant even in the absence of the identified potential risk
factors. The fitted values for all four of the poorly fit cases are quite small.
> fitted(low.reduced)[c(11,13,129,144,152,180)]
11
13
129
144
152
180
0.69818500 0.69818500 0.10930602 0.11486743 0.09877858 0.12307383
Cases 11 and 13 have high predicted probabilities despite the fact that they had babies
with normal birth weight. Their relatively high leverage might come from the fact that
there were very few hypertensive minority women in the study. These two facts
combined lead to the relatively large Cook’s Distances for these two cases.
Plotting Estimated Conditional Probabilities ~ P( Low 1 | x~ )
A summary of the reduced model is given below:
> low.reduced
Call: glm(formula = Low ~ Prev + Hyper + Smoke + Minority + Lwt,
family = binomial)
Coefficients:
(Intercept)
Prev1
-0.26127 1.18194
Hyper1
1.39722
Smoke1
0.98185
Degrees of Freedom: 185 Total (i.e. Null);
Null Deviance:
232.4
Residual Deviance: 200.3
AIC: 212.3
Minority1
1.04480
Lwt
-0.01413
180 Residual
To easily plot probabilities in R we can write a function that takes covariate values and
compute the desired conditional probability.
> x <- seq(min(Lwt),max(Lwt),.5)
>
+
+
+
+
>
+
>
>
>
>
PrLwt <- function(x,Prev,Hyper,Smoke,Minority) {
L <- -.26127 + 1.18194*Prev + 1.39722*Hyper + .98185*Smoke +
1.0448*Minority - .01413*x
exp(L)/(1 + exp(L))
}
plot(x,PrLwt(x,1,1,1,1),xlab="Mother's Weight",ylab="P(Low=1|x)",
ylim=c(0,1),type="l")
title(main="Plot of P(Low=1|X) vs. Mother's Weight")
lines(x,PrLwt(x,0,0,0,0),lty=2,col="red")
lines(x,PrLwt(x,1,1,0,0),lty=3,col="blue")
lines(x,PrLwt(x,0,0,1,1),lty=4,col="green")
67
R Function – Diagplot.log
Plot Cook’s Distance and Delta Deviance for Logistic Regression Models
Diagplot.log = function(glm1)
{
k <- length(glm1$coef)
h <- lm.influence(glm1)$hat
fv <- fitted(glm1)
pr <- resid(glm1, type = "pearson")
dr <- resid(glm1, type = "deviance")
par(mfrow = c(2, 1))
n <- length(fv)
index <- seq(1, n, 1)
Ck <- (1/k)*((pr^2) * h)/((1 - h)^2)
Cd <- dr^2/(1 - h)
plot(index, Ck, type = "n", xlab = "Index", ylab =
"Cook's Distance", cex = 0.7, main =
"Plot of Cook's Distance vs. Index", col = 1)
points(index, Ck, col = 2)
identify(index, Ck)
plot(index, Cd, type = "n", xlab = "Index", ylab =
"Delta Deviance", cex = 0.7, main =
"Plot of Delta Deviance vs. Index")
points(index, Cd, col = 2)
identify(index, Cd)
par(mfrow = c(1, 1))
invisible()
}
68
Interactions and Higher Order Terms (Note ~ uses data frame: Lowbwt)
Working with a slightly different version of the low birth weight data available which
includes an additional predictor, ftv, which is a factor that indicates the number of first
trimester doctor visits the woman (coded as: 0, 1, or 2+). We will examine how the
model below was developed in the next section where we discuss model development.
In the model below we have added an interaction between age and the number of first
trimester visits. The logistic model is:
( x)
~
Age Lwt Smoke Pr ev HT UI
log
o
1
2
3
4
5
6
1 ( x)
~
7 FTV 1 8 FTV 2 9 Age * FTV 1 10 Age * FTV 2 11Smoke * UI
> summary(bigmodel)
Call:
glm(formula = low ~ age + lwt + smoke + ptd + ht + ui + ftv +
age:ftv + smoke:ui, family = binomial)
Deviance Residuals:
Min
1Q
Median
-1.8945 -0.7128 -0.4817
3Q
0.7841
Max
2.3418
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.582389
1.420834 -0.410 0.681885
age
0.075538
0.053945
1.400 0.161428
lwt
-0.020372
0.007488 -2.721 0.006513 **
smoke1
0.780047
0.420043
1.857 0.063302 .
ptd1
1.560304
0.496626
3.142 0.001679 **
ht1
2.065680
0.748330
2.760 0.005773 **
ui1
1.818496
0.666670
2.728 0.006377 **
ftv1
2.921068
2.284093
1.279 0.200941
ftv2+
9.244460
2.650495
3.488 0.000487 ***
age:ftv1
-0.161823
0.096736 -1.673 0.094360 .
age:ftv2+
-0.411011
0.118553 -3.467 0.000527 ***
smoke1:ui1 -1.916644
0.972366 -1.971 0.048711 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 234.67
Residual deviance: 183.07
AIC: 207.07
on 188
on 177
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 4
> bigmodel$coefficients
(Intercept)
age
lwt
smoke1
prev1
ht1
-0.58238913 0.07553844 -0.02037234 0.78004747 1.56030401 2.06567991
ui1
ftv1
ftv2+
age:ftv1
age:ftv2+ smoke1:ui1
1.81849631 2.92106773 9.24445985 -0.16182328 -0.41101103 -1.91664380
69
Calculate P(Low|Age,FTV) for women of average pre-pregnancy weight with all other
risk factors absent. Similar calculations could be done if we wanted to add in other
factors as well.
First we calculate the logits as function of age for three levels of FTV 0, 1, and 2+
respectively.
> L <- -.5824 + .0755*agex - .02037*mean(lwt)
> L1 <- -.5824 + .0755*agex - .02037*mean(lwt) + 2.9211 - .16182*agex
> L2 <- -.5824 + .0755*agex - .02037*mean(lwt) + 9.2445 - .4110*agex
Next we calculate the associated conditional probabilities.
> P <- exp(L)/(1+exp(L))
> P1 <- exp(L1)/(1+exp(L1))
> P2 <- exp(L2)/(1+exp(L2))
Finally we plot the probability curves as function of age and FTV.
> plot(agex,P,type="l",xlab="Age",ylab="P(Low|Age,FTV)",ylim=c(0,1))
> lines(agex,P1,lty=2,col="blue")
> lines(agex,P2,lty=3,col="red")
> title(main="Interaction Between Age and First Trimester
Visits",cex=.6)
The interaction between in age and
FTV produces differences in
direction and magnitude of the age
effect. For women with no first
trimester doctor visits their
probability of low birth weight
increases with age. However for
women with at least one first
trimester visit the probability of low
birth weight decreases with age.
The magnitude of that drop is
largest for women with 2 or more
first trimester visits.
We also have an interaction between smoking and uterine irritability added to the model.
This will affect how we interpret the two in terms of odds ratios. We need to consider
the OR associated with smoking for women without uterine irritability, the OR associated
with uterine irritability for nonsmokers, and finally the OR associated with smoking and
having uterine irritability during pregnancy.
70
These estimated odds ratios are given below:
OR for Smoking with No Uterine Irritability
> exp(.7800)
[1] 2.181472
OR for Uterine Irritability with No Smoking
> exp(1.8185)
[1] 6.162608
OR for Smoking and Uterine Irritability
> exp(.7800+1.8185-1.91664)
[1] 1.977553
This result is hard to explain physiologically and so this interaction term might be
removed from the model.
Model Selection Methods
Stepwise methods used in logistic regression are the same as those used in ordinary least
square regression however the measure is the AIC (Akaike Information Criteria) as
opposed to Mallow’s Ck statistic. Like Mallow’s statistic, AIC balances residual
deviance and the number of parameters in the model.
AIC = D + 2k ˆ
Where D = residual deviance, k = total number of estimated parameters, and ˆ is an
estimate of the dispersion parameter which is taken to be 1 in models where
overdispersion is not present. Overdispersion occurs when the data consists of the
number of successes out of mi > 1 trials and the trials are not independent (e.g. male birth
data from your last homework).
Forward, backward, both forward and backward simultaneously, and all possible subsets
regression methods can be employed to find models with small AIC values. By default R
uses both forward and backward selection simultaneously. The command to do this in R
has the basic form:
> step(current model name)
To have it select from models containing all potential two-way interactions use:
> step(current model name, scope=~.^2)
This sometimes will have problems with convergence due to overfitting (i.e. the
estimated probabilities approach 0 and 1 as in the saturated model). If this occurs you
can have R consider adding each of the potential interaction terms and then you can scan
the list and decide which you might want to add to your existing model. You can then
continue adding terms until the AIC criteria suggests additional terms do not improve
current model.
71
These commands are illustrated for the low birth weight data with first trimester visits
included in the output shown below.
Base Model
> low.glm <- glm(low~age+lwt+race+smoke+ht+ui+ptd+ftv,family=binomial)
> summary(low.glm)
Call:
glm(formula = low ~ age + lwt + race + smoke + ht + ui + ptd +
ftv, family = binomial)
Deviance Residuals:
Min
1Q
Median
-1.7038 -0.8068 -0.5009
3Q
0.8836
Max
2.2151
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.822706
1.240174
0.663 0.50709
age
-0.037220
0.038530 -0.966 0.33404
lwt
-0.015651
0.007048 -2.221 0.02637 *
race2
1.192231
0.534428
2.231 0.02569 *
race3
0.740513
0.459769
1.611 0.10726
smoke1
0.755374
0.423246
1.785 0.07431 .
ht1
1.912974
0.718586
2.662 0.00776 **
ui1
0.680162
0.463464
1.468 0.14222
ptd1
1.343654
0.479409
2.803 0.00507 **
ftv1
-0.436331
0.477792 -0.913 0.36112
ftv2+
0.178939
0.455227
0.393 0.69426
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 234.67
Residual deviance: 195.48
AIC: 217.48
on 188
on 178
degrees of freedom
degrees of freedom
Find “best” model that includes all potential two-way interactions.
> low.step <- step(low.glm,scope=~.^2)
Start: AIC= 217.48
low ~ age + lwt + race + smoke + ht + ui + ptd + ftv
+ age:ftv
- ftv
- age
<none>
- ui
+ smoke:ui
+ lwt:smoke
+ ui:ptd
+ lwt:ui
+ ptd:ftv
+ ht:ptd
Df Deviance
AIC
2
183.00 209.00
2
196.83 214.83
1
196.42 216.42
195.48 217.48
1
197.59 217.59
1
193.76 217.76
1
194.04 218.04
1
194.24 218.24
1
194.28 218.28
2
192.38 218.38
1
194.55 218.55
72
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
age:ptd
age:ht
age:smoke
race:ui
smoke
smoke:ht
smoke:ptd
race
race:smoke
lwt:ptd
lwt:ht
age:lwt
age:ui
ht:ftv
lwt:ftv
smoke:ftv
age:race
lwt:race
race:ptd
lwt
race:ht
ui:ftv
ht
ptd
race:ftv
1
1
1
2
1
1
1
2
2
1
1
1
1
2
2
2
2
2
2
1
2
2
1
1
4
194.58
194.59
194.61
192.63
198.67
195.03
195.16
201.23
193.24
195.35
195.44
195.46
195.47
194.00
194.19
194.47
194.58
194.63
194.83
200.95
195.19
195.32
202.93
203.58
193.81
218.58
218.59
218.61
218.63
218.67
219.03
219.16
219.23
219.24
219.35
219.44
219.46
219.47
220.00
220.19
220.47
220.58
220.63
220.83
220.95
221.19
221.32
222.93
223.58
223.81
Step: AIC= 209
low ~ age + lwt + race + smoke + ht + ui + ptd + ftv + age:ftv
+ smoke:ui
+ lwt:smoke
- race
<none>
+ ui:ptd
+ lwt:ui
+ ht:ptd
- smoke
+ age:smoke
+ race:ui
+ age:ptd
- ui
+ smoke:ht
+ lwt:ptd
+ smoke:ptd
+ age:ht
+ age:ui
+ age:lwt
+ lwt:ht
+ race:smoke
+ lwt:ftv
+ ptd:ftv
+ age:race
+ smoke:ftv
+ ht:ftv
+ lwt:race
+ race:ht
Df Deviance
AIC
1
179.94 207.94
1
180.89 208.89
2
186.99 208.99
183.00 209.00
1
181.42 209.42
1
181.90 209.90
1
182.06 210.06
1
186.11 210.11
1
182.16 210.16
2
180.32 210.32
1
182.50 210.50
1
186.61 210.61
1
182.71 210.71
1
182.75 210.75
1
182.82 210.82
1
182.90 210.90
1
182.96 210.96
1
183.00 211.00
1
183.00 211.00
2
181.23 211.23
2
181.44 211.44
2
181.57 211.57
2
181.62 211.62
2
181.65 211.65
2
181.82 211.82
2
182.55 212.55
2
182.78 212.78
73
+
+
+
-
race:ptd
lwt
ui:ftv
ht
ptd
race:ftv
age:ftv
2
1
2
1
1
4
2
182.85
188.88
182.94
190.13
191.05
181.69
195.48
212.85
212.88
212.94
214.13
215.05
215.69
217.48
Step: AIC= 207.94
low ~ age + lwt + race + smoke + ht + ui + ptd + ftv + age:ftv +
smoke:ui
- race
<none>
+ lwt:smoke
+ ht:ptd
- smoke:ui
+ ui:ptd
+ age:ptd
+ age:smoke
+ smoke:ptd
+ lwt:ptd
+ lwt:ui
+ age:ht
+ smoke:ht
+ age:lwt
+ age:ui
+ lwt:ht
+ lwt:ftv
+ ptd:ftv
+ smoke:ftv
+ race:smoke
+ age:race
+ ht:ftv
+ race:ui
+ ui:ftv
+ race:ht
+ lwt:race
+ race:ptd
- lwt
- ht
+ race:ftv
- ptd
- age:ftv
Df Deviance
AIC
2
183.07 207.07
179.94 207.94
1
178.34 208.34
1
178.89 208.89
1
183.00 209.00
1
179.07 209.07
1
179.35 209.35
1
179.37 209.37
1
179.58 209.58
1
179.61 209.61
1
179.76 209.76
1
179.78 209.78
1
179.82 209.82
1
179.84 209.84
1
179.86 209.86
1
179.94 209.94
2
178.25 210.25
2
178.53 210.53
2
178.64 210.64
2
178.73 210.73
2
178.84 210.84
2
178.89 210.89
2
179.13 211.13
2
179.50 211.50
2
179.52 211.52
2
179.68 211.68
2
179.86 211.86
1
187.15 213.15
1
187.66 213.66
4
178.51 214.51
1
188.83 214.83
2
193.76 217.76
Step: AIC= 207.07
low ~ age + lwt + smoke + ht + ui + ptd + ftv + age:ftv + smoke:ui
<none>
+ lwt:smoke
+ ui:ptd
+ ht:ptd
+ race
+ age:smoke
+ age:ht
Df Deviance
183.07
1
181.40
1
181.88
1
181.93
2
179.94
1
181.97
1
182.64
AIC
207.07
207.40
207.88
207.93
207.94
207.97
208.64
74
+
+
+
+
+
+
+
+
+
+
+
+
+
-
age:ptd
lwt:ptd
lwt:ui
smoke:ptd
age:lwt
smoke:ui
age:ui
smoke:ht
lwt:ht
smoke:ftv
lwt:ftv
ptd:ftv
ui:ftv
ht:ftv
ht
lwt
ptd
age:ftv
1
1
1
1
1
1
1
1
1
2
2
2
2
2
1
1
1
2
182.69
182.73
182.76
182.85
182.92
186.99
182.99
183.02
183.06
181.48
181.69
181.85
182.28
182.41
191.21
191.56
193.59
199.00
208.69
208.73
208.76
208.85
208.92
208.99
208.99
209.02
209.06
209.48
209.69
209.85
210.28
210.41
213.21
213.56
215.59
219.00
Summarize the model returned from the stepwise search
> summary(low.step)
Call:
glm(formula = low ~ age + lwt + smoke + ht + ui + ptd + ftv +
age:ftv + smoke:ui, family = binomial)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.582389
1.420834 -0.410 0.681885
age
0.075538
0.053945
1.400 0.161428
lwt
-0.020372
0.007488 -2.721 0.006513 **
smoke1
0.780047
0.420043
1.857 0.063302 .
ht1
2.065680
0.748330
2.760 0.005773 **
ui1
1.818496
0.666670
2.728 0.006377 **
ptd1
1.560304
0.496626
3.142 0.001679 **
ftv1
2.921068
2.284093
1.279 0.200941
ftv2+
9.244460
2.650495
3.488 0.000487 ***
age:ftv1
-0.161823
0.096736 -1.673 0.094360 .
age:ftv2+
-0.411011
0.118553 -3.467 0.000527 ***
smoke1:ui1 -1.916644
0.972366 -1.971 0.048711 *
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 234.67 on 188 degrees of freedom
Residual deviance: 183.07 on 177 degrees of freedom
AIC: 207.07
Number of Fisher Scoring iterations: 4
This is the model used to demonstrate model interpretation in the presence of
interactions.
An alternative to the full blown search above is to consider adding a single interaction
term to the “Base Model” from the set of all possible terms.
> add1(low.glm,scope=~.^2)
75
Single term additions
Model:
low ~ age + lwt + race
Df Deviance
<none>
195.48
age:lwt
1
195.46
age:race
2
194.58
age:smoke
1
194.61
age:ht
1
194.59
age:ui
1
195.47
age:ptd
1
194.58
age:ftv
2
183.00
lwt:race
2
194.63
lwt:smoke
1
194.04
lwt:ht
1
195.44
lwt:ui
1
194.28
lwt:ptd
1
195.35
lwt:ftv
2
194.19
race:smoke 2
193.24
race:ht
2
195.19
race:ui
2
192.63
race:ptd
2
194.83
race:ftv
4
193.81
smoke:ht
1
195.03
smoke:ui
1
193.76
smoke:ptd
1
195.16
smoke:ftv
2
194.47
ht:ui
0
195.48
ht:ptd
1
194.55
ht:ftv
2
194.00
ui:ptd
1
194.24
ui:ftv
2
195.32
ptd:ftv
2
192.38
+ smoke + ht + ui + ptd + ftv
AIC
217.48
219.46
220.58
218.61
218.59
219.47
218.58
209.00 *
220.63
218.04
219.44
218.28
219.35
220.19
219.24
221.19
218.63
220.83
223.81
219.03
217.76
219.16
220.47
217.48
218.55
220.00
218.24
221.32
218.38
We can than “manually” enter this term to our base model by using the update
command in R.
> low.glm2 <- update(low.glm,.~.+age:ftv)
> summary(low.glm2)
Call:
glm(formula = low ~ age + lwt + race + smoke + ht + ui + ptd +
ftv + age:ftv, family = binomial)
Deviance Residuals:
Min
1Q
Median
-2.0338 -0.7690 -0.4510
3Q
0.8354
Max
2.3383
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.636485
1.558677 -1.050 0.29376
age
0.085461
0.055734
1.533 0.12519
lwt
-0.017599
0.007653 -2.300 0.02147 *
race2
0.994134
0.550962
1.804 0.07118 .
race3
0.700669
0.491400
1.426 0.15391
76
smoke1
0.792972
ht1
1.936204
ui1
0.938620
ptd1
1.373390
ftv1
2.877889
ftv2+
8.264965
age:ftv1
-0.149619
age:ftv2+
-0.359454
--Signif. codes: 0 `***'
0.452303
0.747576
0.492240
0.495738
2.253710
2.594444
0.096342
0.115429
1.753
2.590
1.907
2.770
1.277
3.186
-1.553
-3.114
0.07957
0.00960
0.05654
0.00560
0.20162
0.00144
0.12043
0.00185
.
**
.
**
**
**
0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 234.67 on 188 degrees of freedom
Residual deviance: 183.00 on 176 degrees of freedom
AIC: 209
Number of Fisher Scoring iterations: 4
Next we could use add1 to consider the remaining interaction terms for addition to this
model.
> add1(low.glm2,scope=~.^2)
Single term additions
Model:
low ~ age + lwt + race + smoke + ht + ui + ptd + ftv + age:ftv
Df Deviance
AIC
<none>
183.00 209.00
age:lwt
1
183.00 211.00
age:race
2
181.62 211.62
age:smoke
1
182.16 210.16
age:ht
1
182.90 210.90
age:ui
1
182.96 210.96
age:ptd
1
182.50 210.50
lwt:race
2
182.55 212.55
lwt:smoke
1
180.89 208.89 *
lwt:ht
1
183.00 211.00
lwt:ui
1
181.90 209.90
lwt:ptd
1
182.75 210.75
lwt:ftv
2
181.44 211.44
race:smoke 2
181.23 211.23
race:ht
2
182.78 212.78
race:ui
2
180.32 210.32
race:ptd
2
182.85 212.85
race:ftv
4
181.69 215.69
smoke:ht
1
182.71 210.71
smoke:ui
1
179.94 207.94 **
smoke:ptd
1
182.82 210.82
smoke:ftv
2
181.65 211.65
ht:ui
0
183.00 209.00
ht:ptd
1
182.06 210.06
ht:ftv
2
181.82 211.82
ui:ptd
1
181.42 209.42
ui:ftv
2
182.94 212.94
ptd:ftv
2
181.57 211.57
77
Fitting Logistic Models in Arc and More Diagnostics
from website)
Again we consider the low birth weight case study.
(lowbirtharc.txt
Arc 1.03, rev Aug, 2000, Wed Oct 22, 2003, 12:10:14. Data set name:
Lowbw
Low birth weight study.
Name
Type
n
Info
AGE
Variate 189 Age of mother
BWT
Variate 189 Actual birthweight of child in grams
HT
Variate 189 Mother hypertensive during pregnancy (1 = yes, 0
= no)
ID
Variate 189
LOW
Variate 189 (1 = low birthweight, 0 = normal birthweight)
LWT
Variate 189 Mothers weight at last menstrual cycle
PTD
Variate 189 do not know
PTL
Variate 189 Previous history of premature labor (1 = yes, 0 =
no)
RACE
Variate 189 Race of mother (1 = white, 2 = black, 3 = other)
SMOKE
Variate 189 Mother smoke (1 = yes, 0 = no)
UI
Variate 189 Uterine irritability (1 = yes, 0 = no)
FTV
Text
189 # of doctor visits during 1st trimester
{F}FTV
Factor 189 Factor--first level dropped
{F}HT
Factor 189 Factor--first level dropped
{F}PTD
Factor 189 Factor--first level dropped
{F}RACE Factor 189 Factor--first level dropped
{F}SMOKE Factor 189 Factor--first level dropped
{F}UI
Factor 189 Factor--first level dropped
Select Fit binomial response… from
the Graph & Fit menu
In the resulting dialog box, specify the model as shown on the following page.
78
Give the model a name if you want.
Always include an intercept.
Use the Make Factors… option from the
data set menu to ensure all categorical
predictors are treated as factors.
Put dichotomous response in the
Response… box. The response may also
be the number of “successes” observed.
(see below)
If mi=1 for all cases then put the variable
Ones in the Trials… box. If your
response represented the number of
“successes” observed in mi > 1 trials then
you we need to import the number trials
and put that variable in this box.
The output below shows the results of fitting this initial model.
Data set = Lowbw, Name of Fit = B1
Binomial Regression
Kernel mean function = Logistic
Response
= LOW
Terms
= (AGE LWT {F}FTV {F}HT {F}PTD
Trials
= Ones
Coefficient Estimates
Label
Estimate
Std. Error
Constant
0.386634
1.27736
AGE
-0.0372340
0.0386777
LWT
-0.0156530
0.00707594
{F}FTV[0]
0.436379
0.479161
{F}FTV[2+]
0.615386
0.553104
{F}HT[1]
1.91316
0.720434
{F}PTD[1]
1.34376
0.480445
{F}RACE[2]
1.19241
0.535746
{F}RACE[3]
0.740681
0.461461
{F}SMOKE[1]
0.755525
0.424764
{F}UI[1]
0.680195
0.464216
{F}RACE {F}SMOKE {F}UI)
Est/SE
0.303
-0.963
-2.212
0.911
1.113
2.656
2.797
2.226
1.605
1.779
1.465
p-value
0.7621
0.3357
0.0270
0.3624
0.2659
0.0079
0.0052
0.0260
0.1085
0.0753
0.1429
Note: For FTV those
who went to the
doctor once during
the first trimester are
used as the reference
group
Scale factor:
1.
Number of cases:
189
Degrees of freedom:
178
Pearson X2:
179.059
Deviance:
195.476
(Note: AIC = D + 2k*(scale factor) = 195.48 + 22 = 217.48)
The results are identical those obtained from R.
Null deviance: 234.67
Residual deviance: 195.48
on 188
on 178
degrees of freedom
degrees of freedom
79
Examining Submodels – Backward Elimination and Forward Selection
Forward Elimination –
Select this option and click OK. It will
then show terms are sequentially added
to a model containing any base terms to
the model. By default the base contains
the intercept only.
Backward Elimination –
Simply select this option and click OK. It
will show how terms are sequentially
eliminated from the model along with the
resulting AIC for the deletion.
The other options do what they say.
The results of backward elimination for the current low birth weight model are shown
below.
Data set = Lowbw, Name of Fit = B1
Binomial Regression
Kernel mean function = Logistic
Response
= LOW
Terms
= (AGE LWT {F}FTV {F}HT {F}PTD {F}RACE {F}SMOKE {F}UI)
Trials
= Ones
Backward Elimination: Sequentially remove terms
that give the smallest change in AIC.
All fits include an intercept.
Current terms: (AGE LWT {F}FTV {F}HT {F}PTD {F}RACE {F}SMOKE {F}UI)
df
Deviance
Pearson X2 |
k
AIC
Delete: {F}FTV
180
196.834
180.989
|
9 214.834 *
Delete: AGE
179
196.417
181.401
|
10 216.417
Delete: {F}UI
179
197.585
180.753
|
10 217.585
Delete: {F}SMOKE
179
198.674
186.809
|
10 218.674
Delete: {F}RACE
180
201.227
183.365
|
9 219.227
Delete: LWT
179
200.949
177.855
|
10 220.949
Delete: {F}HT
179
202.934
177.447
|
10 222.934
Delete: {F}PTD
179
203.584
180.74
|
10 223.584
Current terms: (AGE LWT {F}HT {F}PTD {F}RACE {F}SMOKE {F}UI)
df
Deviance
Pearson X2 |
k
AIC
Delete: AGE
181
197.852
183.999
|
8 213.852
Delete: {F}UI
181
199.151
184.559
|
8 215.151
Delete: {F}RACE
182
203.24
182.815
|
7 217.240
Delete: {F}SMOKE
181
201.247
186.953
|
8 217.247
Delete: LWT
181
201.833
181.355
|
8 217.833
Delete: {F}PTD
181
203.948
181.536
|
8 219.948
Delete: {F}HT
181
204.013
179.069
|
8 220.013
*
80
Current terms: (LWT {F}HT {F}PTD {F}RACE {F}SMOKE
df
Deviance
Pearson X2
Delete: {F}UI
182
200.482
186.918
Delete: {F}SMOKE
182
202.567
189.716
Delete: {F}RACE
183
205.466
186.461
Delete: LWT
182
203.816
185.551
Delete: {F}PTD
182
204.217
182.499
Delete: {F}HT
182
205.162
182.282
{F}UI)
|
k
|
7
|
7
|
6
|
7
|
7
|
7
Current terms: (LWT {F}HT {F}PTD {F}RACE {F}SMOKE)
df
Deviance
Pearson X2 |
Delete: {F}SMOKE
183
205.397
189.925
|
Delete: {F}RACE
184
207.955
192.506
|
Delete: {F}HT
183
207.039
184.17
|
Delete: LWT
183
207.165
187.234
|
Delete: {F}PTD
183
208.247
184.45
|
Current terms: (LWT {F}HT {F}PTD {F}RACE)
df
Deviance
Pearson X2 |
Delete: {F}RACE
185
210.123
194.086
|
Delete: {F}HT
184
212.18
188.048
|
Delete: LWT
184
213.226
187.544
|
Delete: {F}PTD
184
216.295
191.533
|
k
6
5
6
6
6
k
4
5
5
5
AIC
214.482
216.567
217.466
217.816
218.217
219.162
*
AIC
217.397
217.955
219.039
219.165
220.247
AIC
218.123
222.180
223.226
226.295
Current terms: (LWT {F}HT {F}PTD)
df
Deviance
Delete: {F}HT
186
217.497
Delete: LWT
186
217.662
Delete: {F}PTD
186
221.142
Pearson X2 |
190.809
|
188.394
|
193.26
|
k
3
3
3
AIC
223.497
223.662
227.142
Current terms: (LWT {F}PTD)
df
Deviance
Delete: LWT
187
221.898
Delete: {F}PTD
187
228.691
Pearson X2 |
188.863
|
189.647
|
k
2
2
AIC
225.898
232.691
* indicates a potential “final” model using the AIC criteria, Arc does not add the *’s.
Making Interactions
To make interactions in Arc…
1st - Select Make
Interactions from
the data set menu.
2nd - Placing all
covariates in the righthand box will create
all possible two-way
interactions.
81
Deciding which interactions to include however is not as easy as in R. You could
potentially include all interactions and then backward eliminate, however things will get
unstable numerically with that many terms in the model. It is better to choose any
interactions you feel might make physiological sense and then backward eliminate.
If Arc does not use the reference group you would like to use, you can create dummy
variables for each level of the factor and then leave the one for the reference group out
when you specify the model.
Selecting these options will
create three dummy
variables one for each level
of FTV (0, 1, 2+).
The model with the age*recoded FTV and the smoking*uterine irritability interactions
we saw in the R handout is summarized below.
Data set = Lowbw, Name of Fit = B6
Binomial Regression
Kernel mean function = Logistic
Response
= LOW
Terms
= (AGE LWT {F}HT {F}PTD {F}SMOKE {F}UI {F}SMOKE*{F}UI
{T}FTV[1] {T}FTV[2+] {T}FTV[1]*AGE {T}FTV[2+]*AGE)
Trials
= Ones
Coefficient Estimates
Label
Estimate
Std. Error
Est/SE
p-value
Constant
-0.582374
1.42158
-0.410
0.6821
AGE
0.0755389
0.0539665
1.400
0.1616
LWT
-0.0203726
0.00749678
-2.718
0.0066
{F}HT[1]
2.06570
0.748727
2.759
0.0058
{F}PTD[1]
1.56032
0.496986
3.140
0.0017
{F}SMOKE[1]
0.780044
0.420371
1.856
0.0635
{F}UI[1]
1.81853
0.667517
2.724
0.0064
{F}SMOKE[1].{F}UI[1] -1.91668
0.973066
-1.970
0.0489
{T}FTV[1]
2.92109
2.28571
1.278
0.2013
{T}FTV[2+]
9.24491
2.66099
3.474
0.0005
{T}FTV[1].AGE
-0.161824
0.0968164
-1.671
0.0946
{T}FTV[2+].AGE
-0.411033
0.119117
-3.451
0.0006
Number of cases:
Degrees of freedom:
Pearson X2:
Deviance:
189
177
179.282
183.073
Notice: The recoding of FTV so FTV=0
is now the reference group.
82
Diagnostic Plots
There are several plotting options in Arc to help assess a models adequacy.
They are as follows:
Residuals (deviance or chi-square) vs. the estimated logit ( Lˆ ˆ T X )
Deviance Residual
y
1 y i
Di 2 sgn( yi ˆ( xi )) yi ln i (1 yi ) ln
ˆ( x )
1 ˆ( x )
i
i
Chi-residual for the ith covariate pattern is defined as:
yi yˆ i
eˆi
the sum of the squared chi-residuals = Pearson’s
m ˆ( x )(1 ˆ( x ))
i
i
~
~i
where yˆ i miˆ( xi ) and yi 1 for cases and 0 for controls.
~
Plot of Cook’s distance vs. Case Number or some other quantity.
Plot of Leverage (potential for influence) vs. Case Number
Model checking plots
Eta’U ~ Estimated Logit ( Lˆi ˆ T xi )
~
Obs-Fraction ~ yi / mi (1 and 0’s in the case mi =1)
ˆ
e Li
Fit-Fraction ~ ˆ( xi )
ˆ
~
1 e Li
Chi-Residuals ~ see above
Dev-Residuals ~ see above
eˆ i
T-Residuals ~
studentized chi-residual
1 hi
Leverages ~ hi = ith element of the hat matrix H
2
1 eˆ i hi
Cook’s Distance ~ Di
measures
k 1 hi 1 hi
influence of the ith case.
Residuals vs. Estimated Logit (or some other function of the covariates)
If the model is adequate, a lowess ( = .6) smooth added to the plot should be constant,
i.e. flat. This plot will not work well when the number of replicates, mi are small, i.e.
close to 1. Model checking plots work better for checking model adequacy in those
cases.
83
As an example consider the simple, but reasonable, main effects model shown on the next
page.
Data set = Lowbw, Name of Fit = B3
Binomial Regression
Kernel mean function = Logistic
Response
= LOW
Terms
= (LWT {F}HT {F}PTD {F}RACE {F}SMOKE {F}UI)
Trials
= Ones
Coefficient Estimates
Label
Estimate
Std. Error
Est/SE
p-value
Constant
-0.125327
0.967238
-0.130
0.8969
LWT
-0.0159185
0.00695085
-2.290
0.0220
{F}HT[1]
1.86689
0.707212
2.640
0.0083
{F}PTD[1]
1.12886
0.450330
2.507
0.0122
{F}RACE[2]
1.30085
0.528349
2.462
0.0138
{F}RACE[3]
0.854413
0.440761
1.938
0.0526
{F}SMOKE[1]
0.866581
0.404341
2.143
0.0321
{F}UI[1]
0.750648
0.458753
1.636
0.1018
Scale factor:
Number of cases:
Degrees of freedom:
Pearson X2:
Deviance:
1.
189
181
183.999
197.852
The plots of the chi-square residuals vs. the estimated logit ( L̂ = ˆ T X ) and LWT are
shown below. The lowess smooth looks fairly flat and so no model inadequacies are
suggested.
84
Cook’s Distance vs. Case Number and Est. Probs - (no cases have high influence)
Leverages vs. Case Numbers
For leverages the average value is k/n, so values far exceeding the average have the
potential to be influential. The following is a good rule of thumb:
1/n < hi < .25 no worries
.25 < hi < .50 worry
.50 < hi < 1 worry lots
Model Checking Plots
For any linear combination b T x i of the predictors of terms imagine drawing two plots:
one of y / m vs. b T x , and one of ˆ( x ) vs. b T x . If the model is adequate lowess smooth
i
i
i
i
~
i
of each should match for any linear combination we choose. A model checking plot is a
plot with b T x i on the x-axis and both the lowess smooths described above added to the
plot. If they agree for a variety of choices of b T x i then we can feel reasonably confident
that our model is adequate. Large differences between these smooths can indicate model
deficiencies. Common choices for b T x i include the estimated logits ( L̂ ), the individual
predictors, and randomly chosen combinations of the terms in the model.
85
Here we see good
agreement between the
two smoothes for the
estimated logits.
Model checking plot
with the single term
LWT on the x-axis.
86
Model checking plot for
one random linear
combination of the terms
in the model. Again we
see good agreement.
87
Using Arc when the Number Trials is not 1
Example 1: Oral contraceptive use, myocardial infarctions, and age
To read these data in Arc it is easiest to create a text file that looks like:
Age OCuse MI NoMI Trials
1 Yes 4 62 66
1 No 2 224 226
2 Yes 9 33 42
2 No 12 390 402
3 Yes 4 26 30
3 No 33 330 363
4 Yes 6 9 15
4 No 65 362 427
5 Yes 6 5 11
5 No 93 301 394
The Trials column contains the total number of patients in each age and oral
contraceptive use category, i.e. the sum of the number of patients with MI and the
number of patients without MI (NoMI).
When read in Arc we have:
; loading D:\Data\Deppa Documents\Biostatistics (Biometry II)\Book
Data\OCMI.txt
Arc 1.06, rev July 2004, Mon Oct 16, 2006, 12:58:46. Data set name:
OCMI
Oral contraceptive use, age, and myocardial infarctions
Name
Type
n
Info
AGE
Variate 10
MI
Variate 10
NOMI
Variate 10
TRIALS Variate 10
OCUSE Text
10
In Arc we need to create to turn the Age variable into a factor as we don’t want to be
interpreted as an actual number and we need to create a factor based on OCuse. By
default Arc does things alphabetically so No would be used as “present” which is not
desirable. Thus it is best to create separate dichotomous dummy variables for each level
individually. This will allow us to use those who used oral contraceptives as having “risk
present”. To do this in Arc we need to use the Make Factors… option in the data menu.
88
For oral contraceptive use we want two separate dummy variables, one for each level of
use, i.e. Yes and No.
Fitting the logistic model in Arc with MI as the response and OCUSE[YES] as the risk
factor indicator.
89
Results for Fitted Logistic Model
Iteration 1: deviance = 6.69914
Iteration 2: deviance = 6.53561
Data set = OCMI, Name of Fit = B1
Binomial Regression
Kernel mean function = Logistic
Response
= MI
Terms
= ({F}AGE {T}OCUSE[YES])
Trials
= TRIALS
Coefficient Estimates
Label
Estimate
Std. Error
Constant
-4.36985
0.434642
{F}AGE[2]
1.13836
0.476782
{F}AGE[3]
1.93440
0.458227
{F}AGE[4]
2.64806
0.449627
{F}AGE[5]
3.19429
0.447386
{T}OCUSE[YES]
1.38518
0.250458
Scale factor:
Number of cases:
Degrees of freedom:
Pearson X2:
Deviance:
Est/SE
-10.054
2.388
4.221
5.889
7.140
5.531
p-value
0.0000
0.0170
0.0000
0.0000
0.0000
0.0000
1.
10
4
6.386
6.536
We can work with these parameter estimates as above to obtain OR’s of interest etc.
90
Motivating Example: Recumbant Cows
“The abiltiy of biochemical and haematolgical tests to predict recovery in
periparturient recumbent cows.” NZ Veterinary Journal, 35, 126-133 Clark, R. G.,
Henderson, H. V., Hoggard, G. K. Ellison, R. S. and Young,B. J. (1987).
Study Description:
For unknown reasons, many pregnant dairy cows become recumbant--they lay down-either shortly before or after calving. This condition can be serious, and may lead to
death of the cow. These data are from a study of blood samples of over 500 cows studied
at the Ruakura (N.Z.) Animal Health Laboratory during 1983-84. A variety of blood
tests were performed, and for many of the animals the outcome (survived, died, or animal
was killed) was determined. The goal is to see if survival can be predicted from the
blood measurements. Case numbers 12607 and 11630 were noted as having exceptional
care---and they survived.
Name
AST
Calving
CK
Daysrec
Inflamat
Myopathy
Outcome
PCV
Urea
CaseNo
Type
Variate
Variate
Variate
Variate
Variate
Variate
Variate
Variate
Variate
Text
n
429
431
413
432
136
222
435
175
266
435
Info
serum asparate amino transferase (U/l at 30C)
0 if measured before calving, 1 if after
Serum creatine phosphokinase (U/l at 30C)
Days recumbent
inflamation 0=no, 1=yes
Muscle disorder, 1 if present, 0 if absent
outcome: 1 if survived, 0 if died or killed (response)
Packed Cell Volume (Haemactocrit), %
serum urea (mmol/l)
case number
Because calving, inflammation, and myopathy are Bernoulli dichotomous predictors they
will not be transformed, although we might consider potential interactions involving
these predictors. We will not consider inflammation and myopathy however as most of
the cows have that information missing.
Guidelines for Transforming Predictors in Logistic Regression
Examining univariate conditional density plots for continuous predictors f ( xi | y )
(Cook & Weisberg)
Consider,
1 if success
f ( x | y ) the conditional density of x given outcome variable y
0 if failure
Idea:
91
Univariate considerations
f ( x | y)
Normal, common variance
i.e. Var ( x | y 0) Var ( x | y 1)
Normal, unequal variances
i.e. Var ( x | y 0) Var ( x | y 1) v
Skewed right
x [0,1]
x is dichotomous, Bernoulli
x ~ Poisson, i.e. x is a count
Suggested model terms
x
x and x2
x and log2(x)
base 2 is easier to interpret
log(x) , log(1-x)
x
x
Multivariate considerations
When considering multiple continuous predictors simultaneously we look at multivariate
normality.
If
f ( x | y ) ~ MVN ( y k , )
~
f ( x | y ) ~ MVN ( y k , y k )
~
then use the x’s themselves
then include x i2 ’s and x i x j terms
For example in the two predictor case (p = 2)
x1 x2 is needed if E ( x1 | x 2 ) o 1, y k x 2
and if the variances are different for the xi across levels of y then we add x i2 terms as
well.
92
AST
Clearly AST has skewed distribution and using the log 2 ( AST ) in the model would be
recommended.
After transformation we have
In f (log 2 ( AST ) | Outcome) appears to approximately normal for both levels with a
constant variance so quadratic terms in the log scale are not suggested.
93
CK
Clearly CK is extremely right skewed and would benefit from log transformation.
Again the conditional densities appear approximately normal with equal variance, so we
will consider adding log 2 (CK ) only to the model.
PCV
f ( PCV | Outcome) is approximately normal for both outcome groups but the variation in
PCV levels appear to be higher for cows that survived. Thus we will consider PCV and
PCV2 terms in the model.
94
Daysrec
Despite the fact that Daysrec is right skewed we will not log transform it. It represents a
count of the number of days the cow was recumbent, so it could be modeled as a Poisson
and thus the only term recommended is the Daysrec itself.
Urea
Consider the log transformation of urea level.
f (log 2 (Urea) | Outcome) is approximately normal however the variation for cows that
survived appears larger so we will consider both log 2 (Urea) and log 2 (Urea) 2 terms.
95
Data set = Downer, Name of Fit = B2
372 cases are missing at least one value. (PCV has lots of missing values also)
Binomial Regression
Kernel mean function = Logistic
Response
= Outcome
Terms
= (AST log2[AST] CK log2[CK] Urea log2[Urea] log2[Urea]^2
PCV PCV^2 Daysrec Calving)
Trials
= Ones
Coefficient Estimates
Label
Estimate
Std. Error
Est/SE
p-value
Constant
-1.03935
6.35298
-0.164
0.8700
AST
-0.000720027
0.00242524
-0.297
0.7666
log2[AST]
-0.330179
0.554239
-0.596
0.5514
CK
-0.000109772
0.000135315
-0.811
0.4172
log2[CK]
-0.0121434
0.223648
-0.054
0.9567
Urea
-1.13453
1.05860
-1.072
0.2838
log2[Urea]
0.730468
2.89371
0.252
0.8007
log2[Urea]^2
0.660165
1.38757
0.476
0.6342
PCV
0.182480
0.224691
0.812
0.4167
PCV^2
-0.00165620
0.00325722
-0.508
0.6111
Daysrec
-0.391937
0.157490
-2.489
0.0128
Calving
1.28561
0.648089
1.984
0.0473
Scale factor:
Number of cases:
Number of cases used:
Degrees of freedom:
Pearson X2:
Deviance:
1.
435
165
153
127.410
141.988
Clearly we have some model reduction to do, as many of the current terms are not
significant. Before backward eliminating we will drop all of the non-transformed
versions of log scale predictors.
Coefficient Estimates
Label
Estimate
Std. Error
Est/SE
Constant
-3.82598
5.84498
-0.655
log2[AST]
-0.554005
0.293416
-1.888
log2[CK]
-0.118575
0.160536
-0.739
log2[Urea]
4.09939
3.12355
1.312
log2[Urea]^2 -0.978895
0.545929
-1.793
PCV
0.218085
0.213730
1.020
PCV^2
-0.00229912
0.00305947
-0.751
Daysrec
-0.383179
0.153758
-2.492
Calving
1.39322
0.647605
2.151
Scale factor:
1.
Number of cases:
435
Number of cases used:
165
Degrees of freedom:
156
Pearson X2:
134.154
Deviance:
145.123
Backward Elimination: Sequentially remove terms
that give the smallest change in AIC.
All fits include an intercept.
p-value
0.5127
0.0590
0.4601
0.1894
0.0730
0.3075
0.4524
0.0127
0.0314
96
Current terms: (log2[AST] log2[CK] log2[Urea] log2[Urea]^2
Daysrec Calving)
df
Deviance
Pearson X2 |
Delete: log2[CK]
157
145.671
134.797
|
Delete: PCV^2
157
145.786
134.995
|
Delete: PCV
157
146.392
135.415
|
Delete: log2[Urea]
157
148.141
140.787
|
Delete: log2[AST]
157
148.92
140.737
|
Delete: Calving
157
150.163
141.672
|
Delete: Daysrec
157
151.993
135.976
|
Delete: log2[Urea]^2
157
152.536
143.299
|
k
8
8
8
8
8
8
8
8
Current terms: (log2[AST] log2[Urea] log2[Urea]^2 PCV
Calving)
df
Deviance
Pearson X2
Delete: PCV^2
158
146.202
135.813
Delete: PCV
158
146.701
136.211
Delete: log2[Urea]
158
149.035
142.035
Delete: Calving
158
151.207
140.587
Delete: Daysrec
158
152.168
136.078
Delete: log2[Urea]^2
158
153.767
145.12
Delete: log2[AST]
158
161.383
144.17
PCV^2 Daysrec
k
7
7
7
7
7
7
7
Current terms: (log2[AST] log2[Urea] log2[Urea]^2 PCV
df
Deviance
Pearson X2
Delete: PCV
159
148.955
137.789
Delete: log2[Urea]
159
150.035
144.626
Delete: Calving
159
152.176
141.179
Delete: Daysrec
159
152.699
136.298
Delete: log2[Urea]^2
159
155.31
149.108
Delete: log2[AST]
159
163.059
140.738
Daysrec
|
k
|
6
|
6
|
6
|
6
|
6
|
6
|
|
|
|
|
|
|
|
PCV PCV^2
AIC
161.671
161.786
162.392
164.141
164.920
166.163
167.993
168.536
AIC
160.202 *
160.701
163.035
165.207
166.168
167.767
175.383
Calving)
AIC
160.955
162.035
164.176
164.699
167.310
175.059
Current terms: (log2[AST] log2[Urea] log2[Urea]^2 Daysrec Calving)
df
Deviance
Pearson X2 |
k
AIC
Delete: log2[Urea]
160
152.373
144.523
|
5 162.373
Delete: Daysrec
160
155.744
138.388
|
5 165.744
Delete: Calving
160
155.99
142.871
|
5 165.990
Delete: log2[Urea]^2
160
157.017
148.417
|
5 167.017
Delete: log2[AST]
160
164.785
143.03
|
5 174.785
Current terms: (log2[AST] log2[Urea]^2 Daysrec Calving)
df
Deviance
Pearson X2 |
Delete: Calving
161
160.932
150.399
|
Delete: Daysrec
161
162.036
146.037
|
Delete: log2[AST]
161
169.755
148.817
|
Delete: log2[Urea]^2
161
176.794
157.24
|
k
4
4
4
4
AIC
168.932
170.036
177.755
184.794
Current terms: (log2[AST] log2[Urea]^2 Daysrec)
df
Deviance
Pearson X2 |
Delete: Daysrec
162
167.184
150.961
|
Delete: log2[AST]
162
178.021
150.618
|
Delete: log2[Urea]^2
162
181.641
162.028
|
k
3
3
3
AIC
173.184
184.021
187.641
Current terms: (log2[AST] log2[Urea]^2)
df
Deviance
Delete: log2[Urea]^2
163
182.688
Delete: log2[AST]
163
192.479
k
2
2
AIC
186.688
196.479
Pearson X2 |
162.386
|
151.943
|
97
Forward selection suggests the same model.
“Final” Model
Data set = Downer, Name of Fit = B5
372 cases are missing at least one value.
Binomial Regression
Kernel mean function = Logistic
Response
= Outcome
Terms
= (log2[AST] log2[Urea] log2[Urea]^2 PCV Daysrec Calving)
Trials
= Ones
Coefficient Estimates
Label
Estimate
Std. Error
Est/SE
p-value
Constant
-1.12404
5.01853
-0.224
0.8228
log2[AST]
-0.733670
0.196044
-3.742
0.0002
log2[Urea]
4.44950
3.17044
1.403
0.1605
log2[Urea]^2 -1.05918
0.554282
-1.911
0.0560
PCV
0.0514512
0.0335256
1.535
0.1249
Daysrec
-0.386695
0.153067
-2.526
0.0115
Calving
1.44641
0.623820
2.319
0.0204
Scale factor:
Number of cases:
Number of cases used:
Degrees of freedom:
Pearson X2:
Deviance:
1.
435
170
163
138.509
148.269
Diagnostics and Model Checking Plots
Chi-residuals vs. estimated logits ~ Looks good.
98
Cook’s Distance and Leverage vs. Case Numbers
Model Checking Plots (Estimated Logits and Marginals)
LOGIT
99
AST
UREA
100
PCV
DAYSREC
All of these plots look OK. The largest departure observed is in the case of urea but the
discrepancy there is primarily due to one observation stands out from the rest.
101
In R
To replicate the analysis above in R you will need the following functions to look and the
conditional densities:
𝑓(𝑥|𝑦 = 0) and 𝑓(𝑥|𝑦 = 1)
The first two functions are used to make pretty histograms in the conplot function. The
function conplot replicates the fitting density estimates conditional on the value of the
outcome variable Y, by taking the predictor X and the Y as arguments. If there are missing
values on either the response or the predictor those cases are automatically removed before
constructing the plot.
nclass.FD = function (x)
{
r <- quantile(x, c(0.25, 0.75))
names(r) <- NULL
h <- 2 * (r[2] - r[1]) * length(x)^{
-1/3
}
ceiling(diff(range(x))/h)
}
bandwidth.nrd = function (x)
{
r <- quantile(x, c(0.25, 0.75))
h <- (r[2] - r[1])/1.34
4 * 1.06 * min(sqrt(var(x)), h) * length(x)^(-1/5)
}
conplot = function (x, xname = deparse(substitute(x)),y)
{
xname <- deparse(substitute(x))
data = na.omit(cbind(x,y))
x = data[,1]
y = data[,2]
par(err = -1)
dens0 <- density(x[y==0], width = bandwidth.nrd(x[y==0]))
dens1 <- density(x[y==1], width = bandwidth.nrd(x[y==1]))
ylim <- range(c(dens0$y,dens1$y))
xlim <- range(c(dens0$x,dens1$x))
hist(x, nclass.FD(x), prob = T, xlab = xname, xlim = xlim,
ylim = ylim,main=paste("Conditional X|Y Plot of ",xname))
lines(dens0,col="blue")
lines(dens1,col="red")
invisible()
}
102
> conplot(x=AST,y=Outcome)
> conplot(x=log(AST),y=Outcome)
Etc…
To obtain model checking plots in R you will need to install the package car from the
CRAN which essentially is a collection of functions to replicate Arc in R. The two
functions that create model checking plots in the car library are called mmp and mmps,
the latter creates model checking plots for each predictor as well as the overall fit.
103
Downer Example in R
> mod1 = glm(Outcome~AST+Urea+PCV+Calving+Daysrec+CK,family="binomial")
> summary(mod1)
Call:
glm(formula = Outcome ~ AST + Urea + PCV + Calving + Daysrec +
CK, family = "binomial")
Deviance Residuals:
Min
1Q
Median
-1.7678 -0.7541 -0.1928
3Q
0.7546
Max
2.0696
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.3313771 1.1987644
0.276 0.78222
AST
-0.0022405 0.0014726 -1.521 0.12815
Urea
-0.3140380 0.0770497 -4.076 4.59e-05 ***
PCV
0.0601745 0.0339726
1.771 0.07652 .
Calving
1.3192777 0.6238318
2.115 0.03445 *
Daysrec
-0.4804961 0.1498000 -3.208 0.00134 **
CK
-0.0001435 0.0001121 -1.280 0.20068
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 212.71 on 164 degrees of freedom
Residual deviance: 146.39 on 158 degrees of freedom
(270 observations deleted due to missingness)
AIC: 160.39
Number of Fisher Scoring iterations: 7
> mmps(mod1)
104
Using the same approach as in the analysis in Arc we might use the model with several
terms based on predictor transformations.
>
>
>
>
>
logAST = log2(AST)
logCK = log2(CK)
logUrea = log2(Urea)
logUrea2 = logUrea^2
PCV2 = PCV^2
> Downer2 = data.frame(Outcome,logAST,logCK,logUrea,logUrea2,PCV,PCV2,Daysrec,Calving)
> Downer2 = na.omit(Downer2)
> attach(Downer2)
> mod2 = glm(Outcome~logAST+logCK+logUrea+logUrea2+PCV+PCV2+
Daysrec+Calving,family="binomial",data=Downer2)
> summary(mod2)
Call:
glm(formula = Outcome ~ logAST + logCK + logUrea + logUrea2 +
PCV + PCV2 + Daysrec + Calving, family = "binomial", data =
Downer2)
Deviance Residuals:
Min
1Q
Median
-1.9522 -0.7094 -0.2869
3Q
0.7109
Max
2.0585
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.826289
5.856671 -0.653
0.5135
logAST
-0.554007
0.293529 -1.887
0.0591 .
logCK
-0.118574
0.160621 -0.738
0.4604
logUrea
4.099642
3.137295
1.307
0.1913
logUrea2
-0.978940
0.548487 -1.785
0.0743 .
PCV
0.218083
0.213771
1.020
0.3076
PCV2
-0.002299
0.003060 -0.751
0.4525
Daysrec
-0.383178
0.153795 -2.491
0.0127 *
Calving
1.393222
0.647759
2.151
0.0315 *
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 212.71
Residual deviance: 145.12
AIC: 163.12
on 164
on 156
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 7
105
Backwards eliminate using the step() function
> mod3 = step(mod2)
Start: AIC=163.12
Outcome ~ logAST + logCK + logUrea + logUrea2 + PCV + PCV2 +
Daysrec + Calving
- logCK
- PCV2
- PCV
<none>
- logUrea
- logAST
- Calving
- Daysrec
- logUrea2
Df Deviance
AIC
1
145.67 161.67
1
145.79 161.79
1
146.39 162.39
145.12 163.12
1
148.14 164.14
1
148.92 164.92
1
150.16 166.16
1
151.99 167.99
1
152.54 168.54
Step: AIC=161.67
Outcome ~ logAST + logUrea + logUrea2 + PCV + PCV2 + Daysrec +
Calving
- PCV2
- PCV
<none>
- logUrea
- Calving
- Daysrec
- logUrea2
- logAST
Df Deviance
AIC
1
146.20 160.20
1
146.70 160.70
145.67 161.67
1
149.03 163.03
1
151.21 165.21
1
152.17 166.17
1
153.77 167.77
1
161.38 175.38
Step: AIC=160.2
Outcome ~ logAST + logUrea + logUrea2 + PCV + Daysrec + Calving
<none>
- PCV
- logUrea
- Calving
- Daysrec
- logUrea2
- logAST
Df Deviance
146.20
1
148.96
1
150.03
1
152.18
1
152.70
1
155.31
1
163.06
AIC
160.20
160.96
162.03
164.18
164.70
167.31
175.06
> summary(mod3)
Call:
glm(formula = Outcome ~ logAST + logUrea + logUrea2 + PCV + Daysrec +
Calving, family = "binomial", data = Downer2)
Deviance Residuals:
Min
1Q
Median
-2.0329 -0.6836 -0.2644
3Q
0.7002
Max
2.0893
106
Coefficients:
Estimate Std. Error z value
(Intercept) -1.49051
5.04944 -0.295
logAST
-0.72986
0.19514 -3.740
logUrea
4.61037
3.19802
1.442
logUrea2
-1.08728
0.55899 -1.945
PCV
0.05489
0.03370
1.629
Daysrec
-0.37191
0.15422 -2.411
Calving
1.45572
0.62845
2.316
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01
Pr(>|z|)
0.767853
0.000184
0.149406
0.051768
0.103394
0.015888
0.020538
***
.
*
*
‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 212.71
Residual deviance: 146.20
AIC: 160.2
on 164
on 158
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 7
> mmps(mod3)
107
We could consider adding interaction terms to our “final” model. This is easily done
using the scope option.
> mod3 = step(mod2,scope=~.^2)
> summary(mod3)
Call:
glm(formula = Outcome ~ logAST + logUrea + logUrea2 + PCV + PCV2
+
Daysrec + Calving + PCV:Calving + logAST:PCV2 + logAST:PCV,
family = "binomial", data = Downer2)
Deviance Residuals:
Min
1Q
Median
-1.8712 -0.6639 -0.1012
3Q
0.6784
Max
2.5298
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 44.950653 39.720672
1.132
0.2578
logAST
-9.472551
5.720701 -1.656
0.0978 .
logUrea
6.107698
3.425871
1.783
0.0746 .
logUrea2
-1.315449
0.602659 -2.183
0.0291 *
PCV
-3.814468
2.386123 -1.599
0.1099
PCV2
0.069737
0.036368
1.918
0.0552 .
Daysrec
-0.388998
0.162716 -2.391
0.0168 *
Calving
12.376954
6.037254
2.050
0.0404 *
PCV:Calving -0.322466
0.168853 -1.910
0.0562 .
logAST:PCV2 -0.010067
0.005179 -1.944
0.0519 .
logAST:PCV
0.608959
0.344917
1.766
0.0775 .
--Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 212.71
Residual deviance: 134.15
AIC: 156.15
on 164
on 154
degrees of freedom
degrees of freedom
Number of Fisher Scoring iterations: 7
108
> mmps(mod3)
There is a slight improvement in fit. The same model fit in JMP produces the following
ROC curve. The resulting classification is very good using this model.
109