Supplementary Material Jiwan et al Contents Pages 1
advertisement

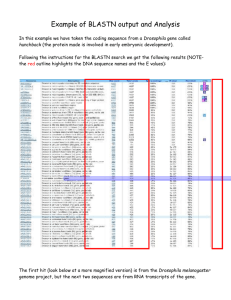
Supplementary Material Jiwan et al Contents Pages 1. Supplementary Table 1 2-10 2. DNA Gel Blot Analysis 11 3. Agrobacterium contamination test 11 4. Alignment of DAJP13 and DAJP14 to MLO homologs in Fragaria vesca 12 1 Supplementary Material Jiwan et al Section 1. Supplementary Table 1. Gene copies included in the phylogenetic analyses of Mlo. Abbreviations for the genes either follow those used previously, or are arbitrarily designated in order to apply unique names to all gene copies in the figures. Species Figure Name GenBank Arabidopsis thaliana AtMlo1 NP_192169 A. thaliana AtMlo2 NP_172598 A. thaliana AtMlo3 NP_566879 A. thaliana AtMlo4 NP_563882 A. thaliana AtMlo5 NP_180923 A. thaliana AtMlo6 NP_176350 A. thaliana AtMlo7 NP_179335 A. thaliana AtMlo8 NP_565416 A. thaliana AtMlo9 NP_174980 A. thaliana AtMlo10 NP_201398 A. thaliana AtMlo11 NP_200187 A. thaliana AtMlo12 NP_565902 A. thaliana AtMlo13 NP_567697 A. thaliana AtMlo14 NP_564257 A. thaliana AtMlo15 NP_181939 Brachypodium distachyon BdMlo1-1 XP_003558907 B. distachyon BdMlo1-2 XP_003558908 B. distachyon BdMlo1-like1 XP_003581276 B. distachyon BdMlo1-like2 XP_003571073 B. distachyon BdMlo4 XP_003577771 B. distachyon BdMlo8 XP_003574266 2 Supplementary Material Jiwan et al B. distachyon BdMlo14 XP_003579492 B. distachyon BdMlo15-1 XP_003575162 Brassica rapa BrMlo1 AAX77014 Capsicum annuum CaMlo1 AAX31277 Chlamydomonas reinhardtii CrMlo XP_001689918 Cucumis melo CmMlo1 ACX55084 Eutrema halophilum EhMlo1 BAJ34386 E. halophilum EhMlo2 BAJ34234 E. halophilum EhMlo3 BAJ33971 E. halophilum EhMlo4 BAJ33805 Fragaria vesca Fv_p09621 F. vesca Fv_p09623 F. vesca Fv_p10531 F. vesca Fv_p12974 F. vesca Fv_p23134 F. vesca Fv_p29691 F. vesca Fv_p31172 Glycine max GmMlo1-1 XP_003522288 G. max GmMlo1-2 XP_003527594 G. max GmMlo1-3 XP_003547751 G. max GmMlo1-4 XP_003523525 G. max GmMlo1-5 XP_003520359 G. max GmMlo3-1 XP_003520647 G. max GmMlo3-2 XP_003553544 G. max GmMlo4-1 XP_003538881 3 Supplementary Material Jiwan et al G. max GmMlo4-2 XP_003516545 G. max GmMlo5-1 XP_003523524 G. max GmMlo5-2 XP_003527633 G. max GmMlo6-1 XP_003555433 G. max GmMlo6-2 XP_003540745 G. max GmMlo8-1 XP_003517123 G. max GmMlo8-2 XP_003517124 G. max GmMlo8-3 XP_003548777 G. max GmMlo8-4 XP_003534232 G. max GmMlo11-1 XP_003526073 G. max GmMlo11-2 XP_003543248 G. max GmMlo12-1 XP_003526251 G. max GmMlo12-2 XP_003548913 G. max GmMlo12-3 XP_003548912 G. max GmMlo12-4 XP_003540949 G. max GmMlo13 XP_003546228 Hordeum vulgare HvMlo1 P93766 H. vulgare HvMlo2 BAJ97456 H. vulgare HvMlo3 AAS93431 H. vulgare HvMlo4 BAK00279 H. vulgare HvMlo5 BAK08220 Lotus japonicus LjMlo1 AAX77015 Malus domestica Md_p119 M. domestica Md_p123 M. domestica Md_p141 4 Supplementary Material Jiwan et al M. domestica Md_p145 M. domestica Md_p146 M. domestica Md_p163 M. domestica Md_p168 M. domestica Md_p191 M. domestica Md_p196 M. domestica Md_p207 M. domestica Md_p239 M. domestica Md_p928 Malus toringoides MtMlo1 ADV29809 Medicago truncatula MtrMlo1 ADV40949 M. truncatula MtrMlo2 XP_003612195 M. truncatula MtrMlo3 XP_003604071 M. truncatula MtrMlo4 XP_003603856 M. truncatula MtrMlo5 XP_003594424 M. truncatula MtrMlo6 XP_003592974 M. truncatula MtrMlo7 XP_003607521 M. truncatula MtrMlo8 XP_003636206 M. truncatula MtrMlo9 XP_003636497 M. truncatula MtrMlo10 XP_003636499 M. truncatula MtrMlo11 XP_003612408 M. truncatula MtrMlo12 XP_003603854 M. truncatula MtrMlo14 XP_003629098 Oryza alta OaMlo ACN85277 O. australiensis OauMlo ACN85283 5 Supplementary Material Jiwan et al O. coarctata OcMlo ACN85293 O. granulata OgMlo ACN85338 O. nivara OnMlo ACN85176 O. officinalis OoMlo ACN85251 O. ridleyi OrMlo ACN85306 O. sativa OsMlo1 A2YD22 O. sativa OsMlo2 AAK94907 O. sativa OsMlo3 AAK72963 Ostreococcus lucimarinus OlMlo XP_001420898 Physcomitrella patens subsp. patens PpaMlo1 XP_001768083 P. patens subsp. patens PpaMlo2 XP_001784403 P. patens subsp. patens PpaMlo3 XP_001781175 P. patens subsp. patens PpaMlo4 XP_001776768 P. patens subsp. patens PpaMlo5 XP_001770178 P. patens subsp. patens PpaMlo6 XP_001763298 P. patens subsp. patens PpaMlo7 XP_001763297 P. patens subsp. patens PpaMlo8 XP_001754053 Pisum sativum PsMlo1 ACO07297 Populus trichocarpa PtMlo1 XP_002310696 P. trichocarpa PtMlo2 XP_002307201 P. trichocarpa PtMlo3 XP_002306989 P. trichocarpa PtMlo4 XP_002314276 P. trichocarpa PtMlo5 XP_002301907 P. trichocarpa PtMlo6 XP_002301905 6 Supplementary Material Jiwan et al P. trichocarpa PtMlo7 XP_002327651 P. trichocarpa PtMlo8 XP_002331489 P. trichocarpa PtMlo9 XP_002319210 P. trichocarpa PtMlo10 XP_002310695 P. trichocarpa PtMlo11 XP_002309708 P. trichocarpa PtMlo12 XP_002312021 P. trichocarpa PtMlo13 XP_002315295 Populus trichocarpa x P. deltoides PtxdMlo ABK96389 Prunus americana PaMlo1 ACZ81392 P. americana PaMlo2 ACZ81391 P. persica PpMlo1 ACZ81390 P. persica PpMlo2 P. persica PpMlo3 P. persica PpMlo4 P. persica PpMlo5 P. persica PpMlo6 P. persica PpMlo7 P. persica PpMlo8 P. persica PpMlo9 P. persica PpMlo10 P. persica PpMlo11 P. persica PpMlo12 P. persica PpMlo13 P. persica PpMlo14 7 Supplementary Material Jiwan et al P. persica PpMlo15 P. persica PpMlo16 Ricinus communis RcMlo1 XP_002533335 R. communis RcMlo2 XP_002510710 R. communis RcMlo3 XP_002510708 R. communis RcMlo4 XP_002517839 R. communis RcMlo5 XP_002522497 R. communis RcMlo6 XP_002526372 R. communis RcMlo7 XP_002526370 R. communis RcMlo8 XP_002533246 R. communis RcMlo9 XP_002532472 R. communis RcMlo10 XP_002530672 R. communis RcMlo11 XP_002528562 Selaginella moellendorffii SmMlo1 XP_002961667 S. moellendorffii SmMlo2 XP_002963507 S. moellendorffii SmMlo3 XP_002965989 S. moellendorffii SmMlo4 XP_002965637 S. moellendorffii SmMlo5 XP_002967927 S. moellendorffii SmMlo6 XP_002971112 S. moellendorffii SmMlo7 XP_002972783 S. moellendorffii SmMlo8 XP_002981181 Solanum lycopersicum SlMlo1 NP_001234814 Sorghum bicolor SbMlo1 XP_002438667 S. bicolor SbMlo2 XP_002465008 S. bicolor SbMlo3 XP_002447339 8 Supplementary Material Jiwan et al S. bicolor SbMlo4 XP_002441108 S. bicolor SbMlo5 XP_002447908 S. bicolor SbMlo6 XP_002439069 S. bicolor SbMlo7 XP_002441107 S. bicolor SbMlo8 XP_002449108 S. bicolor SbMlo9 XP_002454019 S. bicolor SbMlo10 XP_002465934 Triticum aestivum TaMlo1 AAS93630 T. aestivum TaMlo2 AAK94904 T. aestivum TaMlo3 BAJ24148 T. aestivum TaMlo4 BAJ24149 T. aestivum TaMlo5 BAJ24150 T. aestivum TaMlo6 BAJ24151 T. aestivum TaMlo7 BAJ24153 Vitis vinifera VvMlo CAN84002 V. vinifera VvMlo1-like XP_002273002 V. vinifera VvMlo4 XP_002266927 V. vinifera VvMlo5 XP_002266377 V. vinifera VvMlo5-like XP_002273026 V. vinifera VvMlo6 XP_002274608 V. vinifera VvMlo6-like XP_002273434 V. vinifera VvMlo7 XP_002274642 V. vinifera VvMlo9 XP_002276608 V. vinifera VvMlo10 XP_002275360 V. vinifera VvMlo11 XP_002275390 9 Supplementary Material Jiwan et al V. vinifera VvMlo12 XP_002282198 V. vinifera VvMlo13 XP_002282216 V. vinifera VvMlo14 XP_002282190 V. vinifera VvMlo15 XP_002280697 V. vinifera VvMlo16 XP_002266144 V. vinifera VvMlo17 XP_002265520 Zea mays ZmMlo1 NP_001105660 Z. mays ZmMlo2 NP_001105168 Z. mays ZmMlo3 NP_001105527 Z. mays ZmMlo4 NP_001105169 Z. mays ZmMlo5 ACN34145 Z. mays ZmMlo6 NP_001105170 Z. mays ZmMlo7 NP_001105661 Z. mays ZmMlo8 NP_001105171 10 Supplementary Material Jiwan et al 2. Section 2 – DNA Gel Blot Analysis Gel blot analysis was performed using standard protocols as described earlier (Sambrook et al., 2000). Briefly, 10 g of total DNA was digested with BglII restriction enzyme. DNA was electrophoresed on a 0.8% agarose gel. Prior to transfer to a membrane, the gel was depurinated, denautured and neutralized with prescribed solutions. DNA was transferred to Nylon membrane and crosslinked using a UV crosslinker (Stratagene Inc.). DNA was prehybridized and hybridized with a PpMLO1 gene probe as described previously (Dhingra et al., 2004). Hybridized blot was washed and processed for autoradiography. Expected fragments of 2.35 kb in Fa-pDAJ3 and 2.86 kb in Fa-pDAJ4 along with different sized higher fragments confirmed that these transgenics represented different transgenic events. Supplementary Figure 1 References: Sambrook J, MacCallum P, Russell D (2000) Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor Laboratory Press, NY Dhingra A, Portis AR, Daniell H (2004) Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proceedings of the National Academy of Sciences of the United States of America 101 (16):6315-6320 11 Supplementary Material Jiwan et al 3. Section 3 - Agrobacterium contamination test During Agrobacterium-mediated transformation, there is a possibility of bacterial contamination in the regenerated plants. The PCR results detailed above could be due to the presence of plasmid DNA within the contaminating bacteria. To eliminate the possibility of bacterial contamination, we used PCR with virB1 gene specific primers DAJP 11 and DAJP12 (Table 1). As shown in Supplementary Figure 2, positive amplification of a 300 bp region nested in the virB1 gene is only observed in the AGL0 host bacterial strain which was used at a very high dilution for PCR. Absence of the amplicon in samples labeled as Fa-pDAJ3.1-3.3 and Fa-pDAJ4.1-4.3 indicates the absence of Agrobacterium contamination in transgenic lines. Supplementary Figure 2 Figure 2 – Assessment of Agrobacterium contamination Primers annealing to virB1, DAJP11 and DAJP12 were used to test for presence of Agrobacterium. An amplicon of 300 bp was obtained when AGL0 strain was used as a template. The amplicon is missing in Fa-pDAJ 3.1-3.3 and Fa-pDAJ4.1-4.3 lines. Lane marked as M represents DNA ladder. 4. Section 4 - Alignment of DAJP13 and DAJP14 to MLO homologs in Fragaria vesca Table shows the location and percentage identity of primers with MLO homologs predicted in Fragaria vesca. DAJP13 and DAJP14 are predicted to generate a 953 bp amplicon in Fragaria vesca with cDNA as a template. Note the complementarity at the GC-rich 3’ end of the primers. Gene DAJP13 DAJP14 10995 12974 (PpMLO1 Homolog) 686-708 1060-1083 130-157 %Identity DAJP13/DAJP14 79/79/71 Amplicon size 953 bp 12 Supplementary Material 29691 31172 09621 09623 10134 10531 28466 23134 1051-1074 394-417 613-634 1547-1570 1330-1358 172-195 1385-1408 - Jiwan et al 806-831 115-144 1477-1503 79/75/67/64 67/66/62/60 62/-/61 Sequence alignment of DAJP13 and DAJP14 with gene12974 a PpMLO1 homolog predicted to be present in Fragaria vesca genome. Note the similarity at 3’end that is GC rich. Query 28nt >DAJP13 Target 1674nt >gene12974 Qry Tgt 1 + AAAGTTGGAACGTTCCTCACGGATAGGC 28 | ||| ||||||| || | || |||| 130 + ACAGTCGGAACGTGGCTTTCAGAGAGGC 157 28 cols, 20 ids (71.4%), 0 gaps (0.0%) Query 24nt >DAJP14 Target 1674nt >gene12974 Qry 24 - CGGCCTCAGTTGGTTCTTCATTTT 1 || || || |||||||||||| | Tgt 1060 + CGACCGCACTTGGTTCTTCATCTC 1083 24 cols, 19 ids (79.2%), 0 gaps (0.0%) 13