EPH_1354_sm_supmat
advertisement

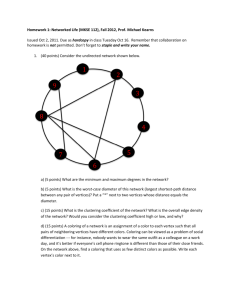
The influence of gap junction network complexity on pulmonary artery smooth muscle reactivity in normoxic and chronic hypoxic conditions Marko Gosak, Christelle Guibert, Marie Billaud, Etienne Roux and Marko Marhl Supplemental material – intercellular network model The network is generated on the bases of the so-called vertex fitness network model (Caldarelli et al., 2002; Servedio et al., 2004) and the spatially embedded vertex fitness network model (Morita, 2006; Gosak et al., 2012). First, we randomly distribute N C vertices in a rectangular box with dimensions 1.0×1.0×0.1 with uniform distribution. In order to ensure a certain degree of homogeneity in the positioning of the vertices, we include a minimal allowed distance ( lmin 0.08 ) between the vertices. Then, to each vertex a fitness value f i is prescribed, which are assumed to follow a power law distribution P( f ) ~ f . Fitness values are assigned deterministically as follows: f i (i/N )1 /(1 ) , (S1) where i 1,..., N C and 2.5 is the scaling exponent. The condition to link vertex i with vertex j is: fi f j (li , j ) , (S2) where li , j signifies the Euclidean distance between i -th and j -th vertex, is the control parameter characterizing the topology of the network and is the threshold. If the left term in Eq. S2 exceeds the threshold, then the i -th and j -th vertex are connected with each other. By choosing a proper we can set the mean vertex degree k (i.e. the average number of connections of individual cells) in the network. In other words, for each construction of the 1 network we numerically determine the desired value , for which the desired mean degree of the network is attained. For 1, where the connectivity is predominantly dependent on the fitness of the vertices, long-range interactions are present in the network structure (Gosak et al., 2012). However, as 1, the Euclidean distance becomes the key constrain that defines the topology. For 1.8 we have a rather heterogeneous network with a high tendency of neighboring vertices to be connected with each other. Furthermore, for even higher values of (such as 5 ), only the Euclidean distance impacts the connectivity. The resulting network is thus a very homogeneous random geometric network. Network structures obtained with 1.8 and 5 are regarded as possible candidates for models for the cytoarchitecture of the intrapulmonary artery and are shown in Fig. S1. Basic metrics for the exploration of complex structural properties in networks are the average path length and the clustering coefficient. The average path length L is a measure of functional integration and defines the average number of mediating links along the shortest path between any two nodes, whereas the clustering coefficient – a measure of functional segregation – is related to the cliquishness of a typical neighborhood in the network and is defined as the number of existing connections between all neighbors of a node divided by the number of all possible connections between them. In particular, if the node degree of the i-th vertex is denoted by ki, there are ki(ki-1)/2 possible links between its neighbors. The clustering coefficient of the i-th node Ci is then given by the fraction of those links that are actually present in the graph. The network’s average clustering coefficient Cavg is estimated by simply averaging Ci over all the vertices. Calculation of the shortest path length can be problematic in networks with disconnected vertices where the distance between two nodes may be infinite. For that reason the global efficiency E is commonly introduced to reflect the traffic capacity of a network, and is defined as follows: E i j d ij1 N C ( N C 1) (A3) , where d ij is the length of the shortest path from unit i to unit j. Notably, E is inversely related to the average shortest path length L. To characterize the topological features of the network model we show in Fig. S2 the standard deviation of vertex degrees k (reflecting the level of heterogeneity of the network), the average clustering coefficient and the average shortest path length as a function of the network parameter . It can be observed that indeed the network obtained by 1.8 is rather heterogeneous, efficient and highly clustered, whereas the 2 network generated by 5 is homogeneous, less efficient and exhibits a lower clustering coefficient. Notably, the network obtained by 1.8 simultaneously display both high integration and segregation, i.e. high efficiency and clustering, which is a feature of smallworld networks (Watts&Strogatz, 1998). Figure S1. 3D representation of the network for 1.8 (A) and 5 (B). In both cases the mean node degree was set to k 6 and the number of cells was NC=200, with the z coordinate 10-fold smaller than the x, y coordinates. Figure S2. Standard deviation of node degrees (A), the average clustering coefficient (B) and the average shortest path length (C) as a function of . 3 References Caldarelli G, Capocci A, DeLosRios P & Muñoz MA (2002). Scale-free networks from varying vertex intrinsic fitness. Phys Rev Lett 89, 258702. Gosak M, Markovič R & Marhl M (2012). The role of neural architecture and the speed of signal propagation in the process of synchronization of bursting neurons. Physica A 391, 2764–2770. Morita S (2006). Crossovers in scale-free networks on geographical space. Phys Rev E 73 035104(R). Servedio VDP, Calderelli G & Buttà P (2004). Vertex intrinsic fitness: How to produce arbitrary scale-free network. Phys Rev E 70, 056126. Watts DJ & Strogatz SH (1998). Collective dynamics of 'small-world' networks. Nature 393, 440-442. 4