review questions - biology4friends
advertisement

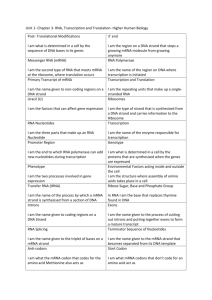
7.3 Transcription 1. What is the difference between sense and antisense strands in DNA? The antisense strand is complementary to the sense strand. The antisense strand is also known as the template strand, as it is used for transcription, to make mRNA. mRNA corresponds with the coding DNA strand, with the exception of the base U, which is replaced with T. mRNA is complementary to the antisense strand 2. What is the role of the antisense strand in transcription? DNA has two complementary strands and only one of these strands is transcribed into RNA, which is the antisense strand. During this process the antisense strand is being transcribed into RNA, having a sequence that will be complementary to the RNA sequence. This will be the “DNA version” of the tRNA anticodon sequence. 3. What is the direction of transcription? Transcription is carried out in the 5’--- 3’ direction 4. What are the roles of the following regions of DNA in transcription? Promoter region: starts the initiation of transcription where the RNA polymerase attaches so that the DNA can be unwound Coding region: transcribed section of DNA Terminator: serves to end transcription 5. Explain the process of transcription in prokaryotes, including the following: promoter region, RNA polymerase, 5’-3’ direction, free nucleoside triphosphates, complementary base pairing, terminator region. The process of transcription in prokaryotes begins when the RNA polymerase binds to the promoter, which causes the unwinding and separation in the DNA strands. Then the nucleoside triphosphates bind to the complementary bases on the antisense strand pairing the adenine with the uracil, and the cytosine binds with the guanine. The RNA polymerase synthesizes a RNA strand in a 5’—3’ direction until it reaches the terminator. At the terminator the RNA polymerase and the newly formed RNA strand both detach from the antisense template and the DNA unwinds. 1. State one method of post-transcription modification of mRNA in eukaryotes. One method of post-transcription modification of mRNA in eukaryotes is RNA slicing. The In pre-mRNAs there are regions that do not code for translated protein which are called introns which can interrupt the coding regions known as exons. Therefore these areas must be removed. As soon as the pre-mRNAs is transcribed, it is bound by snRNPs, which are responsible for splicing introns out of pre-mRNAs. snRNPs bind to consensus sequences and the snRNPs contain RNA molecules that can bond through complementary base pairing to the consensus sequences. The snRNPs come together to forma spliceosome and as this spliceosome forms the intron loops out. By cutting the pre-mRNA at one intron-exon boundary, where it leaves reactive free hydroxyl group on the exon. The spliceosome uses this hydroxyl group to attack the other end of the intron, in order to remove the introns entirely. Then once the introns are removed, the exons are able to join together, forming a mature mRNA molecule. 2. Describe the genetic code: (starts on translation) - Molecule = sequence of bases on mRNA - Function= tells ribosome what aa (amino acid) to use - Codon = set of 3 bases on mRNA, recognized by anticodon on tRNA - Start codon = 3 letter code for the first aa- same for nearly all polypeptides always AUG, translates to methionine/ MET - Stop codon =3 letter code that follows the last aa- says “put no aa here” 6. Compare transcription and translation Transcription Translation Begins with… DNA mRNA Ends with… RNA Polypeptide Location Nucleus Cytoplasm Uses… RNA polymerase Ribosome