Table S5. Selected genes affected in condition EL
advertisement
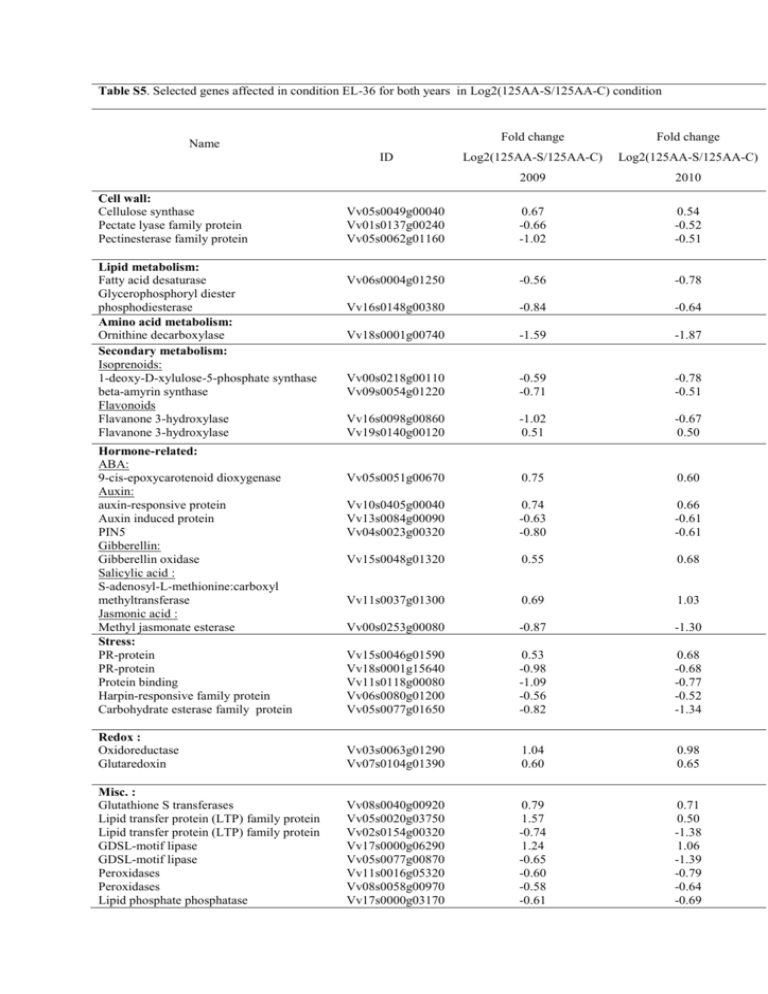
Table S5. Selected genes affected in condition EL-36 for both years in Log2(125AA-S/125AA-C) condition Fold change Fold change Log2(125AA-S/125AA-C) Log2(125AA-S/125AA-C) 2009 2010 Vv05s0049g00040 Vv01s0137g00240 Vv05s0062g01160 0.67 -0.66 -1.02 0.54 -0.52 -0.51 Vv06s0004g01250 -0.56 -0.78 Vv16s0148g00380 -0.84 -0.64 Vv18s0001g00740 -1.59 -1.87 Vv00s0218g00110 Vv09s0054g01220 -0.59 -0.71 -0.78 -0.51 Vv16s0098g00860 Vv19s0140g00120 -1.02 0.51 -0.67 0.50 Vv05s0051g00670 0.75 0.60 Vv10s0405g00040 Vv13s0084g00090 Vv04s0023g00320 0.74 -0.63 -0.80 0.66 -0.61 -0.61 Vv15s0048g01320 0.55 0.68 Vv11s0037g01300 0.69 1.03 Vv00s0253g00080 -0.87 -1.30 Vv15s0046g01590 Vv18s0001g15640 Vv11s0118g00080 Vv06s0080g01200 Vv05s0077g01650 0.53 -0.98 -1.09 -0.56 -0.82 0.68 -0.68 -0.77 -0.52 -1.34 Redox : Oxidoreductase Glutaredoxin Vv03s0063g01290 Vv07s0104g01390 1.04 0.60 0.98 0.65 Misc. : Glutathione S transferases Lipid transfer protein (LTP) family protein Lipid transfer protein (LTP) family protein GDSL-motif lipase GDSL-motif lipase Peroxidases Peroxidases Lipid phosphate phosphatase Vv08s0040g00920 Vv05s0020g03750 Vv02s0154g00320 Vv17s0000g06290 Vv05s0077g00870 Vv11s0016g05320 Vv08s0058g00970 Vv17s0000g03170 0.79 1.57 -0.74 1.24 -0.65 -0.60 -0.58 -0.61 0.71 0.50 -1.38 1.06 -1.39 -0.79 -0.64 -0.69 Name ID Cell wall: Cellulose synthase Pectate lyase family protein Pectinesterase family protein Lipid metabolism: Fatty acid desaturase Glycerophosphoryl diester phosphodiesterase Amino acid metabolism: Ornithine decarboxylase Secondary metabolism: Isoprenoids: 1-deoxy-D-xylulose-5-phosphate synthase beta-amyrin synthase Flavonoids Flavanone 3-hydroxylase Flavanone 3-hydroxylase Hormone-related: ABA: 9-cis-epoxycarotenoid dioxygenase Auxin: auxin-responsive protein Auxin induced protein PIN5 Gibberellin: Gibberellin oxidase Salicylic acid : S-adenosyl-L-methionine:carboxyl methyltransferase Jasmonic acid : Methyl jasmonate esterase Stress: PR-protein PR-protein Protein binding Harpin-responsive family protein Carbohydrate esterase family protein Transcription: MYB family transcription factor MYB family transcription factor MYB family transcription factor MYB family transcription factor Homeobox transcription factor CCAAT box binding factor MADS box transcription factor WRKY transcription factor family WRKY transcription factor family Aux/IAA family NIM1-INTERACTING ARF transcription factor family Signalling: Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Protein kinase family protein Calmodulin Calmodulin Calcium ion binding Development: LEA Transport: Amino acid transporter Lysine/histidine transporter Proton-dependent oligopeptide transport Peptide/nitrate transporter Carnitine transporter Sugar transporter Bidirectional sugar transporter SWEET Bidirectional sugar transporter SWEET Carbohydrate/Phosphate transmembrane transporter Carbohydrate transmembrane transporter Sodium transporter PLEIOTROPIC DRUG RESISTANCE Vv07s0197g00060 Vv02s0033g00450 Vv07s0005g01950 Vv01s0026g01910 Vv18s0001g10160 Vv00s0956g00020 Vv18s0041g01880 Vv00s2547g00010 Vv07s0031g01710 Vv14s0030g02310 Vv17s0000g03780 Vv11s0016g03540 0.66 0.58 0.54 -0.59 0.52 -0.65 -0.53 -0.52 -0.91 -0.59 -1.16 -0.68 0.81 0.53 0.80 -0.60 0.82 -0.59 -1.04 -0.53 -0.62 -0.56 -1.01 -0.73 Vv01s0026g01420 Vv16s0050g01960 Vv19s0027g00010 Vv04s0008g00320 Vv00s2588g00010 Vv00s0258g00090 Vv16s0148g00100 Vv00s2254g00010 Vv00s1467g00010 Vv14s0068g00100 Vv00s0424g00020 Vv11s0118g00050 Vv18s0001g10630 Vv04s0023g01100 Vv05s0049g01780 0.56 1.15 -0.70 -0.88 -0.94 -0.65 -0.83 -0.93 -0.67 -1.44 -0.63 -0.59 0.62 -0.94 1.40 0.68 0.92 -0.54 -0.91 -0.55 -0.54 -0.59 -0.57 -0.62 -0.77 -0.50 -0.65 1.23 -0.54 1.08 Vv08s0007g04240 1.11 1.70 Vv18s0001g03540 Vv01s0011g03180 Vv18s0041g00560 Vv08s0007g02020 Vv19s0085g00920 Vv05s0020g02170 Vv07s0104g01340 Vv14s0066g01420 -0.67 -0.52 -0.53 -0.89 -0.77 1.02 0.71 -0.57 -0.90 -0.57 -2.18 -0.57 -1.17 1.39 0.70 -0.85 Vv08s0007g01810 Vv00s0304g00020 Vv11s0103g00010 Vv06s0061g01480 0.68 -0.68 -0.58 -0.57 1.06 -0.51 -1.66 -0.51