Megan Roosen-Runge October 8, 2010 PHG 519 Homework 1 In a
advertisement
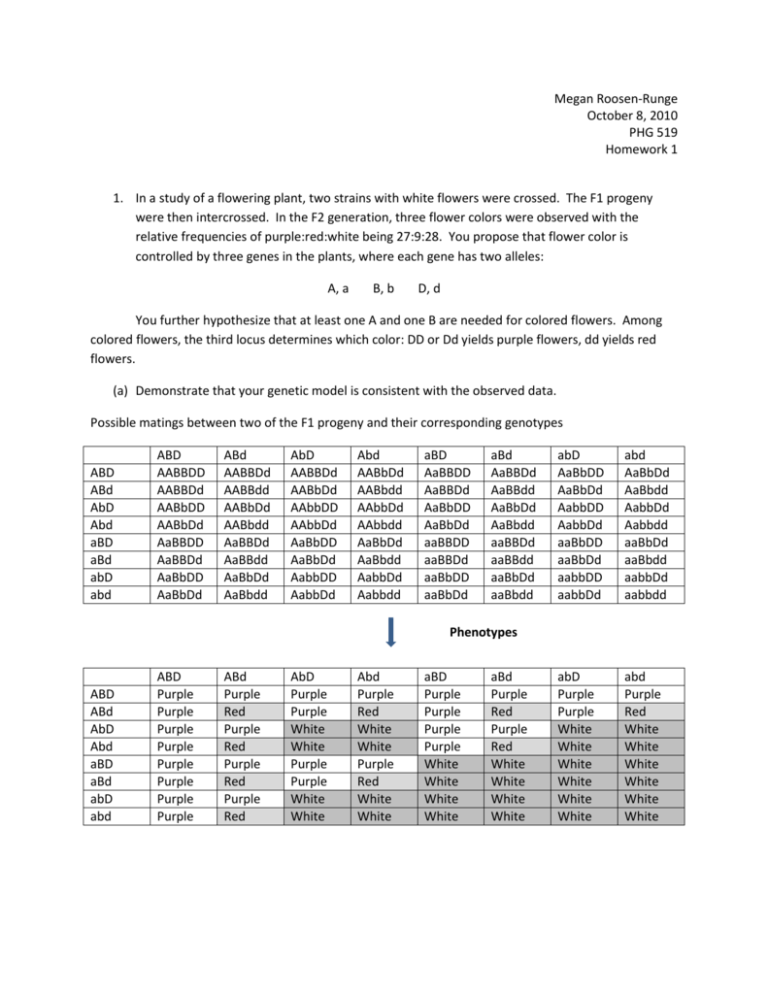
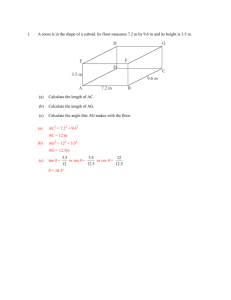
Megan Roosen-Runge October 8, 2010 PHG 519 Homework 1 1. In a study of a flowering plant, two strains with white flowers were crossed. The F1 progeny were then intercrossed. In the F2 generation, three flower colors were observed with the relative frequencies of purple:red:white being 27:9:28. You propose that flower color is controlled by three genes in the plants, where each gene has two alleles: A, a B, b D, d You further hypothesize that at least one A and one B are needed for colored flowers. Among colored flowers, the third locus determines which color: DD or Dd yields purple flowers, dd yields red flowers. (a) Demonstrate that your genetic model is consistent with the observed data. Possible matings between two of the F1 progeny and their corresponding genotypes ABD ABd AbD Abd aBD aBd abD abd ABD AABBDD AABBDd AABbDD AABbDd AaBBDD AaBBDd AaBbDD AaBbDd ABd AABBDd AABBdd AABbDd AABbdd AaBBDd AaBBdd AaBbDd AaBbdd AbD AABBDd AABbDd AAbbDD AAbbDd AaBbDD AaBbDd AabbDD AabbDd Abd AABbDd AABbdd AAbbDd AAbbdd AaBbDd AaBbdd AabbDd Aabbdd aBD AaBBDD AaBBDd AaBbDD AaBbDd aaBBDD aaBBDd aaBbDD aaBbDd aBd AaBBDd AaBBdd AaBbDd AaBbdd aaBBDd aaBBdd aaBbDd aaBbdd abD AaBbDD AaBbDd AabbDD AabbDd aaBbDD aaBbDd aabbDD aabbDd abd AaBbDd AaBbdd AabbDd Aabbdd aaBbDd aaBbdd aabbDd aabbdd abD Purple Purple White White White White White White abd Purple Red White White White White White White Phenotypes ABD ABd AbD Abd aBD aBd abD abd ABD Purple Purple Purple Purple Purple Purple Purple Purple ABd Purple Red Purple Red Purple Red Purple Red AbD Purple Purple White White Purple Purple White White Abd Purple Red White White Purple Red White White aBD Purple Purple Purple Purple White White White White aBd Purple Red Purple Red White White White White As shown in the phenotype table above, intercrossing two heterozygote F1 progeny with the genotype AaBbDd yields 64 genotype possibilities in the F2 generation. Of those F2 progeny, we observe the correct phenotypic ratio 27 purple: 9 red: 28 white. (b) What color(s) were the flowers in F1? With what relative frequencies? As mentioned in the description of this problem, the parental generation consisted of two plants with white flowers. Since the parents in any Mendelian model are obligated to be homozygotes and our F1 generation is obligated to be all heterozygotes, our cross looked like this: AAbbDD X aaBBdd Parental generation AaBbDd F1 generation Thus, all of the F1 flowers are purple, since there is one A and one B allele and the D locus is not “dd.” Since all progeny in the F1 generation have the same genotype, the relative frequency is 100% purple. Note: Other parental genotype possibilities are: AAbbdd and aaBBDD. (c) Consider a backcross of F1 plants to one of the parental strains. What color are the flowers and with what relative frequency? Backcross: AAbbDD x AaBbDd Possible matings and the resulting genotypes of their progeny: AbD ABD AABbDD ABd AABbDd Abd AAbbDd AbD AAbbDD aBD AaBbDD abD AabbDD aBd AaBbDd abd AabbDd Abd White AbD White aBD Purple abD White aBd Purple abd White Corresponding phenotypes: AbD ABD Purple ABd Purple Based on the above table, we would expect to see a 1:1 (50%, 50%), purple:white ratio amongst the progeny resulting from the backcross of AAbbDD (parental) and AaBbDd (F1). (d) Consider the backcross of F1 plants to the other parental strain. What color are the flowers and with what relative frequency? Backcross: aaBBdd x AaBbDd Possible matings and the resulting genotypes of their progeny: ABD AaBBDd aBd ABd AaBBdd Abd AaBbdd AbD AaBbDd aBD aaBBDd abD aaBbDd aBd aaBBdd abd aaBbdd AbD Purple aBD White abD White aBd White abd White (e) Corresponding phenotypes: aBd ABD Purple ABd Red Abd Red Based on the above table, we would expect to see a 1:1:2 (25%, 25%, 50%), purple:red: white ratio amongst the progeny resulting from the backcross of aaBBdd (parental) and AaBbDd (F1). 2) In humans, assume that the presence of freckles is an autosomal dominant trait controlled by a single gene. Suppose that in a particular family, both parents have freckles. The couple has a daughter that has freckles and a son that does not have freckles. What are the genotypes of the four people in the family? The couple is expecting a new addition to the family in December 2010. What is the probability that their third child will be born without freckles? What is the probability that their third child will be born with freckles. Gene: F, f alleles Mother’s genotype: Ff Father’s Genotype: Ff Daughter’s genotype: F? Son’s Genotype: ff For the third child: there is a 75% chance their child will have freckles (25% chance of FF + 50% chance of Ff) and there is a 25% chance their child will not have freckles. 3) In humans, assume that having attached earlobes is controlled by a single gene. Having attached earlobes is recessive to having free earlobes. A woman and her husband both have free earlobes, although each happens to have a father with attached earlobes. Is it possible for this woman and man to have a child with attached earlobes? Explain by indicating the genotypes of the parents and child. ee ee Ee Ee ? It is possible for this couple to have a child with attached earlobes (ee). Because the parents are heterozygotes for this trait, their offspring will have the genotype ratio of 1:2:1 (EE:Ee:ee), which gives a 25% chance that their child will have attached earlobes. 4) Cockayne’s syndrome is a rare disease. 100 families with one affected parent were recruited for a genetic study of this disease. In the sample, there were a total of 167 offspring, of which 70 are affected and 97 are unaffected. You would like to determine if the observed data is consistent with an autosomal dominant mode of inheritance for Cockayne’s syndrome. (a) Give the appropriate null hypothesis H0 and alternative hypothesis Ha for testing whether Cockayne’s syndrome is an autosomal dominant disease. H0: p=1/2 vs. Ha: p≠1/2 (this assumes that there are no affected homozygotes, since most rare diseases are lethal in the homozygote form) (b) Calculate your test statistic and the corresponding P-value for your test statistic. X=# affected = 70; n= 167 X ~ B(167, ½ ) Since np≥10 and n(1-p)≥10 (167*0.5=83.5), we can use the normal approximation to the binomial. For this study of Cockayne’s syndrome, with 70 affected offspring, we get the following: Z= 70−167(0.5) =-2.089 √(167∗0.5)(0.5) The p-value is 2P(Z≥|𝑧|)=2(0.0184)=0.0368 (c) What is your conclusion about the validity of an autosomal dominant model for the disorder? Based on this data concerning Cockayne’s syndrome, we found a statistically significant result at the 95% confidence level (P=0.0368). Thus, this data and its corresponding statistical evidence do not support an autosomal dominant model of inheritance. That model is inconsistent with this syndrome. 5) Assume there is a diploid organism that has 7 pairs of chromosomes. Suppose the organism is heterozygous at only one locus on each of these seven pairs of chromosomes (Aa, Bb, …, Gg). Independent assortment will permit the production of how many genetically different gametes? There are 27 possible gametes, given only one heterozygous locus on each of these seven chromosomes. In other words, this situation yields a total of 128 genetically different gametes.