Additional file 1: Supplemental methods
advertisement
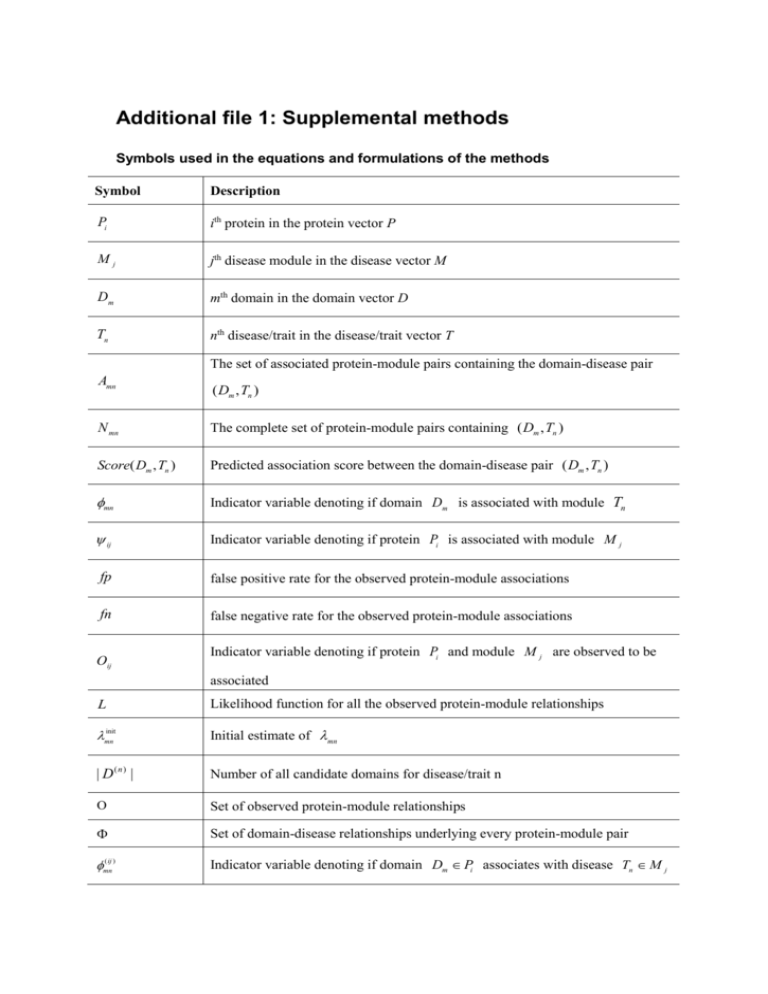
Additional file 1: Supplemental methods
Symbols used in the equations and formulations of the methods
Symbol
Description
Pi
ith protein in the protein vector P
Mj
jth disease module in the disease vector M
Dm
mth domain in the domain vector D
Tn
nth disease/trait in the disease/trait vector T
The set of associated protein-module pairs containing the domain-disease pair
Amn
( Dm , Tn )
N mn
The complete set of protein-module pairs containing ( Dm , Tn )
Score( Dm , Tn )
Predicted association score between the domain-disease pair ( Dm , Tn )
mn
Indicator variable denoting if domain Dm is associated with module Tn
ij
Indicator variable denoting if protein Pi is associated with module M j
fp
false positive rate for the observed protein-module associations
fn
false negative rate for the observed protein-module associations
Oij
Indicator variable denoting if protein Pi and module M j are observed to be
associated
L
Likelihood function for all the observed protein-module relationships
init
mn
Initial estimate of mn
| D(n) |
Number of all candidate domains for disease/trait n
Set of observed protein-module relationships
Set of domain-disease relationships underlying every protein-module pair
( ij )
mn
Indicator variable denoting if domain Dm Pi associates with disease Tn M j
X mn
Number of associated ( Dm , Tn ) domain-disease pairs in the associated
protein-module pairs
Number of non-associated ( Dm , Tn ) domain-disease pairs in the associated
Ymn
protein-module pairs
Set of non-associated protein-module pairs containing the domain-disease pair
Z mn
a,
( Dm , Tn )
b
Predefined integers used in the DPEA approach
Obtained from by setting the probability of domain Dm associated with
mn
disease Tn to be 0
u p , v p , un , v n , ,
Hyper-parameters used in the Bayesian approach
hij ( )
Identical to Pr ( ij 1)
Set of domain-disease pairs underlying all associated protein-module pairs
Variable to measure the strength of potential domain-disease association between
xmn
r
domain Dm and disease Tn .
Reliability rate, the probability that a protein-module association actually exists
The average xmn obtained by including each protein-module association into
LP-score
the constraints with probability r and performing the linear programming
1,000 times
Number of occurrences (witnesses) for a given domain-disease pair ( Dm , Tn ) in
w( Dm , Tn )
each associated protein-module pair ( Pi , M j )
Frequency of obtaining the same or higher LP-score in the 1,000 runs when the
p-value( Dm , Tn )
pw-score
protein-module pair containing the domain-disease pair ( Dm , Tn ) is randomized
Promiscuity versus witnesses (pw)-score to each domain-disease pair
Maximum Likelihood Estimation (MLE) approach
From the main text, the probability of protein Pi associating with module M j is
Pr ( ij 1) 1
( Dm ,Tn )( Pi , M j )
(1 mn )
(1)
The probability for the observed protein-module association is
Pr(Oij 1) Pr(Oij 1, ij 1) Pr(Oij 1, ij 0)
Pr(Oij 1 | ij 1) Pr( ij 1) Pr(Oij 1 | ij 0)(1 Pr( ij 1))
(2)
Pr( ij 1)(1 fn) (1 Pr( ij 1)) fp
Then the likelihood function is
L (Pr(Oij 1)) ij (1 Pr(Oij 1))
1Oij
O
(3)
ij
which is a function of { , fp, fn} .
Therefore we define the complete data as (, ) , in which {Oij oij , i j} is the
( ij )
, Dm Pi , Tn M j } is set
set of observed protein-module relationships, and {mn
( ij )
1
of domain-disease relationships underlying every protein-module pair, where mn
if domain Dm associates with disease/trait Tn in the protein-module pair ( Pi , M j )
( ij )
0 otherwise. We derive the forms of the EM algorithm as follows.
and mn
E-step:
E (
( ij )
mn
M-step:
| Oij oij ,
( t 1)
)
( ij )
Pr(mn
1, Oij oij | ( t 1) )
Pr(Oij oij | ( t 1) )
( ij )
( ij )
Pr(mn
1 | ( t 1) ) Pr(Oij oij | mn
1, ( t 1) )
Pr(Oij oij | ( t 1) )
( ij )
Pr(mn
1 | ( t 1) ) Pr(Oij oij | Pij 1, ( t 1) )
Pr(Oij oij | ( t 1) )
1Oij
( t 1)
mn
(1 fn ) fn
Pr(Oij oij | ( t 1) )
Oij
(t )
mn
1
N mn
Dm Pi , Tn M j
( ij )
E (mn
| Oij oij , ( t 1) )
(4)
The EM algorithm is implemented as follows:
(n)
Step 1. Initialize parameters {mn , fp, fn} as {1/ | D |,0, 0.9} , and compute
Pr( ij 1) by Equation (1) and Pr(Oij 1) by Equation (2);
Step 2. Update parameter {mn } by Equation (4) and compute the likelihood function
L by Equation (3);
Step 3. Go to Step 2, repeat until the value of L is unchanged (within certain error, in
this paper we use 1e-5).
Domain-disease pair exclusion analysis (DPEA) approach
( ij )
In order to deduce the score function of the DPEA approach, we define mn
as the
indicator variable denoting if domain Dm Pi associates with disease Tn M j . For
( ij )
( ij )
simplicity we initialize all mn
1 . In addition, we also define X mn mn as the
ij
number of associated ( Dm , Tn ) domain-disease pairs in the associated
( ij )
protein-module pairs, Ymn (1 mn ) as the number of non-associated ( Dm , Tn )
ij
domain-disease pairs in the associated protein-module pairs, and
Z mn {( P, M ); Dm P, Tn M , P is not associated with M } as the set of
non-associated protein-module pairs containing the domain-disease pair ( Dm , Tn ) .
Let the initial estimate of mn be
init
mn
X mn
X mn
Ymn Z mn
The likelihood L of the observed protein-module associations is estimated as
L mn
X mn a
1 mn
Ymn Zmn b
mn
Here a and b are predefined integers to prevent mn from being exactly 0 or 1 in
the case of few occurrences of ( Dm , Tn ) pairs in the data, and thus extremely high or
low mn can arise only from large numbers of observations pertaining to the
potential association of ( Dm , Tn ) . In our study, both a and b are set to be 1, as
was used by Riley et al. [14].
The likelihood L is a function of mn , and we apply an EM algorithm to
estimate . Next, instead of using mn , we use the change in log-likelihood of
observed protein-module associations as a score to measure the strength of association
for the domain-disease pair ( Dm , Tn ) , when ( Dm , Tn ) is assumed to be not
associated. The score is thus defined as
Score( Dm , Tn ) log
ij
1
log
ij
Pr(Oij 1 | Dm , Tn can associate)
Pr(Oij 1 | Dm , Tn do not associate)
(1 kl )
1
( Dk ,Tl )( Pi , M j )
1
( Dk ,Tl )( Pi , M j )
where
mn
mn
kl
is obtained from by setting the probability of domain Dm
associated with disease Tn to be zero, and is also estimated by the EM algorithm.