Supplementary for A simple method to estimate the average
advertisement

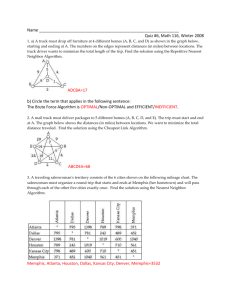
Supplementary for A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment Ulrike Endesfelder, Sebastian Malkusch, Franziska Fricke, Mike Heilemann Institute of Physical and Theoretical Chemistry, Johann Wolfgang Goethe University Frankfurt am Main, Frankfurt am Main, Germany Corresponding authors: Ulrike Endesfelder (endesfelder@chemie.uni-frankfurt.de, +49-69-798-29425, fax +49-69-798-29560) Mike Heilemann (Heilemann@chemie.uni-frankfurt.de, +49-69-798-29736, fax +49-69-798-29560) Monte Carlo simulations of fluorophore distributions to characterize NN adfr In order to investigate the influence of false nearest neighbors on the distribution for a single molecule with Gaussian distributed localizations, we simulated fluorophore distributions with distances beyond the diffraction limit (~200 nm) and defined structures. First, a linear structure labeled with fluorophores was simulated containing five fluorophores that are placed in equal distances along one axis (supplementary figure 2, three representative distances). The simulation reveals periodical Gaussianshaped contributions with declining amplitude in the nearest neighbor distribution NNadfr, each possessing a center of mass exactly at the simulated distance and its multiples (e.g. xc1 = 60 nm, xc2 = 120 nm, xc3 = 180 nm for a simulated distance of 60 nm between two fluorophores supplementary fig.2b). They directly reflect the labeling density of the simulated linear structure. The decrease in amplitude is explained by the nature of the nearest neighbor measurement: In case of two localizations in the same frame, which is getting more likely for increasing distances dependent on experimental conditions and algorithmic performances, the nearest neighbor approach favors the closer localization. To simulate a more densely labeled two-dimensional structure, a single cell of a rhombic Bravais lattice with five fluorophores placed in a face centered square was chosen. The simulation results in a single Gaussian-shaped appearance in the distance distribution NN adfr centering between the distance of the vertex and the center and the distance of two adjacent vertexes (supplementary figure 3). Taking the result for the linear simulation into account, the Gaussian-shaped appearance is defined by three overlapping Gaussian distributions, each indicating one of the three distances (vertex to vertex at the same side, vertex to center and diagonal distance of vertex to vertex) inside the lattice. Based on the results of these two simulations we postulate that the short-range order disturbance depends, as expected, on the structure and the labeling density of the sample. It is reasonably well approximated by a Gaussian distribution reflecting these factors. As for real experiments the sample and labeling properties are usually not known, we add a Gaussian correction term for short range order with free fit parameters and estimated initial values to our distribution equation p(d). For distances above ~ 100 nm, the nearest neighbor distance distribution NNadfr shows a constant population of marginal amplitude that is not explained by the equation, correcting for short-range order disturbance. We define this population as long-range order disturbance, as its significance emerges with increasing distances, while the structural effect on the distribution NNadfr is getting smaller (see 1 linear simulation). Notably, as the area scales quadratic with the distance, the detection of a fluorophore within a distinct distance interval becomes more likely for larger distances. We simulated a uniform distribution of localizations with a single fluorophore in the center (supplementary figure 4). We obtain a linear shape with positive slope and an origin at zero contributing to the distance distribution NNadfr. We defined the influence of the long-range order disturbance with a linear factor in p(d). The gradient is depending on the structure and the degree of labeling, which typically are not known in practice. It is important to mention that p(d) is only approximating false nearest neighbor distributions at distances of about the diffraction limit. For larger distances, another correction term weighting the linear part is needed, as in case two or more molecules are fluorescent, the nearest neighbor selection would always yield the shortest distance to the closest molecule. For a SMLM experiment only one other molecule should reside in the fluorescent state in an adjacent frame and the range of the diffraction-limited area. Thus there is no need for further correction terms that increase the parameter range and would make the fitting error prone. 2 Supplementary Figure 1: Monte Carlo simulation of individual blinking molecule. By the repeated simulation (100 individual simulations per condition) of the distribution of localization of an individual molecule ( = 10 nm, number of localizations 100, 200, 300 and 900) the precision by measuring the Gaussian distribution of the absolute distances to the center of mass (red box plots) and the corresponding relative distribution of the pairwise distances of all localizations (black box 𝑛𝑙𝑜𝑐 −1 plots) is verified. The box plot reveals the higher statistics of the pairwise distribution (𝑛 = ∑𝑖=1 (𝑖)) in contrast to the statistics of the absolute distance measure (𝑛 = 𝑛𝑙𝑜𝑐 , 𝑛𝑙𝑜𝑐 number of localizations) and the spread of localization precision which enhances significantly with the number of localizations (box: 25th to 75th percentile, square: median, line: mean, bars: 5th and 95th percentile, crosses: 1th and 99th percentile, minus: minimum and maximum). 3 Supplementary Figure 2: Monte Carlo simulation of five blinking molecules in a row. By simulating several molecules in a row, the influence on the nearest neighbor distribution of localizations in adjacent frames for one-dimensional structures (e.g. filaments) is evaluated (each molecule 3000 localizations, on = 1 frame). The contribution is quantified by calculating the difference spectrum of the nearest neighbor displacement measure of all localizations of the simulation (middle panel, blue curve) in respect to the nearest neighbor displacement measure of the middle molecule only (middle panel, green curve) and is depicted in the right panel. The maxima of the interfering distributions reflect the simulated distance and its multiples of the molecules. 4 Supplementary Figure 3: Monte Carlo simulation of five blinking molecules in 2D structure. By simulating five molecules arranged in a 2D cross, the influence on the nearest neighbor distribution of localizations in adjacent frames for two-dimensional structures is evaluated (each molecule 3000 localizations, on = 1 frame). The contribution is quantified by calculating the difference spectrum of the nearest neighbor displacement measure of all localizations of the simulation (middle panel, blue curve) in respect to the nearest neighbor displacement measure of the middle molecule only (middle panel, green curve) and is depicted in the right panel. In general, the interfering distribution of distances between different molecules can be described as a sum of all non-Gaussian distributions of pairwise nearest neighbor distances from the different molecules which are in close proximity. This analytical distribution nevertheless can only be described by knowing the number of molecules marking the specific structure and the ability to assign each individual localization to its corresponding molecule. As in a typical localization microscopy experiment localization distributions of molecules overlap, for small distances, nearest neighbors belonging to the same molecule cannot be distinguished from those involving localizations from different molecules. As seen in the maxima of the interfering contribution (right panel) for different distances between the simulated molecules, they give a measure of the average labeling density of the 2D structure. 5 Supplementary Figure 4: Monte Carlo simulation of a blinking molecule and uniform background disturbance. By simulating uniform background (red localizations), the influence of background on the nearest neighbor distribution of localizations in adjacent frames is evaluated (molecule blue, 3000 localizations, on = 1 frame). As seen in the progression of (a-c), the contribution of nearest neighbors of different molecules increases with background density. The contribution is quantified by calculating the difference spectrum of the nearest neighbor displacement measure of all localizations of the simulation (middle panel, blue curve) in respect to the nearest neighbor displacement measure of the molecule only (middle panel, green curve) and is depicted in the right panel. As the density of possible nearest neighbor localizations from the background increases radically from the localization of interest, an increasing contribution is obtained. 6