sibship technique
advertisement

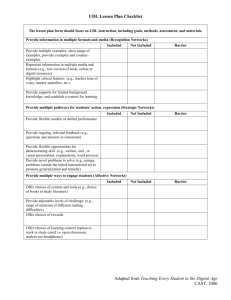
Summary of Sibship Method of Determining Young-of-year passage through Crossings By Jason Coombs, Post-doc research assistant, Univ of Massachusetts, Amherst June 2010 Barriers restricting, or even eliminating, individual movement among freshwater habitat patches has become a common occurrence. This reduction, or loss, of movement potential results in population fragmentation, which in turn increases rates of population inbreeding and extirpation. Thus, barrier remediation within freshwater systems has become a priority for both conservationists and managers alike. However, barrier remediation efforts face two key challenges. The first involves ranking the overwhelming number of potential barriers in freshwater systems to determine which remediation effort would provide the greatest impact to affected populations. The second challenge is determining if the remediation effort was successful. Both of these challenges require addressing the basic question “Are individuals moving across the barrier, and if so in which direction?”. Traditionally, this would require the use of a capture-mark-recapture (CMR) experimental design. Outlined briefly, individuals would be initially captured, marked with an individual or habitat specific identifier, and returned before executing a second capture event after an elapsed period of time to recapture marked individuals and assess movement rates and directions. However, the advent and advancement of molecular techniques and relationship reconstruction algorithms has made another alternative possible. Like CMR, the populations adjacent to the barrier would be sampled. Unlike CMR, individuals would not be tagged, but instead donate a tissue sample as a source of DNA. Molecular techniques would then acquire individual genotypes which, when run through a relationship reconstruction algorithm, would delineate individuals into full-sibling families. Movement across a barrier would then be determined by whether all members of a full-sibling family were on one side or both sides of the barrier. Directionality of movement could be attained through use of a majority-rule approach, where the side with the greatest proportion of family members is assumed to be the natal patch, and direction of movement would be away from this side. In addition to this key assumption, a second assumes that a parental pair reproduces on only one side of a barrier. Initial data for brook trout (Salvelinus fontinalis) shows these assumptions to be likely, but requires further assessment. Simulations assessing the accuracy of this method for a variety of population differentiation (DST = 0, 0.1, 0.25) and movement restriction (two-way, one-way, none) scenarios have been positive, with greater than 95% accuracy for assigned movers and directionality achieved for all scenarios. Additionally, the use of this method does not preclude the use of traditional CMR techniques in that individual genotypes can be used as individual identifiers. Furthermore, unlike traditional CMR techniques, this method is able to use information on parent locations during spawning/birth to determine natal patch origin, lessening the dependence on the majority-rule assumption. Parents can either be known directly or assigned genetically through relationship reconstruction algorithms. In summary, we feel that this method holds great promise for barrier assessment in that it is able to provide estimates of both movement rate and directionality without the time lag of traditional CMR techniques.