pce12067-sup-0005-si

SUPPORTING INFORMATION
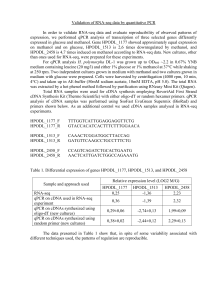
Table S1. Primers used in this study.
All primers were synthesized by Eurofins MWG Operon (www.operon.com).
Name
Sequence (5’ to 3’)
Purpose gi1cds+1F gi1cds+3489R
GIcds+1F
GIcds+3522R
GIpromoter-2310F
GIpromoter-1R
CACCATGTCAGATTCAAATGTGAAGTGGA
TTGATGGG
TCATGACGGGAGAGGGCAGC
CACCATGGCTAGTTCATCTTCATCTGAGA
GATG
TTATTGGGACAAGGATATAGTACAGCCG
CACCACCAGCATATCTCTAATCAG
GATGCGGCCGCACCGAAACTAAACCCCA
AC
GRMZM5G816228F CGA GAC CAG GAG AAC CAA AAG
Cloning
Cloning
Cloning
Cloning gi1 gi1
GI
GI
CDS
CDS
CDS
CDS
Cloning GI promoter
Cloning GI promoter qPCR normalization for maize
GRMZM5G816228R TGA GGA GTT TGT CCA TAA CCA C conz1qF conz1qR zcn8qF zcn8qR gi1qF gi1qR
PP2AqF
PP2AqR qPCR normalization for maize
GGTCAGTGCTTACACAGATTC
GAGCTTGGCATGTCTGTC
TACTTCTGCCAAGTGCTGAGCTA
TGTCACTGCCACCGATATCG
CCA TTC CTG GAT CTG ACA TCG qPCR qPCR qPCR qPCR
ATT AGA GCC GTC CTA CTC GC qPCR of native transcript qPCR of native transcript
TAT CGG ATG ACG ATT CTT CGT GCA G qPCR normalization for
Arabidopsis
GCT TGG TCG ACT ATC GGA ATG AGA G qPCR normalization for
Arabidopsis
AtGIqF
AtGIqR gi1q2F gi1q2R
GGGTAAATATGCTGCTGGAGA
CAGTATGACACCAGCTCCATT
TGA TGT CGT TTC AGC CTC AC
CAA GGC TGC AAG TCC TTC TC qPCR of GI qPCR of GI qPCR of GIp::gi1 lines qPCR of GIp::gi1 lines
SUPPORTING INFORMATION FIGURE LEGENDS
Figure S1. Characterization of GI and gi1 expression in transgenic Arabidopsis lines carrying either the GIp::gi1 or GIp::GI constructs in the gi-201 background.
Expression of gi1 (A) and GI (B) in WT, gi-201 , and two homozygous T3 lines each for
GIp::gi1 and GIp::GI at 8 hours after dawn (ZT8). Above each graph is a schematic of the corresponding transcriptional fusion. Plants were grown in LD photoperiods for 1 week. Expression of gi1 and GI in these samples was measured as the number of transcript molecules present in a 2
μL aliquot of diluted first-strand cDNA (see “Materials and Methods”).
PP2A served as normalization standard (Shin, Park & Choi, 2007).
The number of transcript molecules in samples was calculated from a standard curve based on Ct value from qPCR reactions
(see “Materials and Methods”). To account for technical variation, gi1 and GI expression is presented as the ratio of their transcript number to that of PP2A . Data are the mean of three experimental replicates and error bars are standard deviation.
Figure S2. Expression of gi1 in the gi1-m1::Mu3 and gi1-m2::Mu1 mutant alleles
compared to the A632 inbred. gi1 transcript levels in A632 (closed squares), gi1m1::Mu3 (open circles) and gi1-m2::Mu1 (open triangles) at the indicated time points after dawn (time 0 hours (h)) in leaf samples from V6 stage plants determined with qPCR using the primers in Table SI. Plants were maintained in SD photoperiods in a growth chamber with a 3:1 mix of cool white fluorescent bulbs and incandescent bulbs.
Data are the mean of three experimental replicates and error bars are standard deviation. The grey region denotes the dark period of the photoperiod.
Figure S3. Flowering phenotype of gi1 mutant alleles in the BC1F3 generation.
The total number of leaves produced by A632 (black bar; n = 113), gi1-m1::Mu3 (grey bar; n = 67), gi1-m2::Mu1 (white bar; n = 49), and gi1-m1::Mu3 X gi1-m2::Mu1 F1 hybrid
(F1; diagonal striped bar; n = 26) plants were counted in field grown plants from three trials planted summer of 2011. The F1 hybrid seed was from two separate crosses.
Each trial consisted of 10 to 20 plants planted at 8-day intervals beginning in early June.
Leaf number is the average of counts from the three trials and error bars are standard deviation. Asterisks over gi1-m1::Mu3 , gi1-m2::Mu1 , and gi1-m1::Mu3 X gi1-m2::Mu1 F1 indicate p-values of <0.001 (***) when compared to A632 with an unpaired two-tailed ttest.
Figure S4. miR156 and miR172 accumulated to normal levels in gi1 mutant alleles. miR172 and miR156 levels were determined for A632, gi1-m1::Mu3 , and gi1-m2::M1 in whole seedlings at the V3 stage by northern blot with 32 P-labeled oligonucleotide probes complementary to either zma-MIR156 and zma-MIR172c. The same blot was probed separately for each microRNA. Blots were performed as described previously (Chuck et al., 2007, Park et al., 2002). Labeled arrows indicate the position of the band corresponding to either miR172 or miR156.