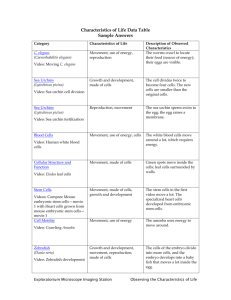
Plant Pathogens Research Paper
advertisement

Methanol and Aqueous Extract of Terminalia bellirica.: A Quorum Sensing Inhibitor and the Novel Application of Terminalia bellirica as an Environmental and Cost-Effective Measure to Inhibit Plant Pathogens in Crops. A Plant Science and Environmental Paper Presented to Junior Science, Engineering, and Humanities Symposium Seattle Pacific University by Indira Rayala Junior Henry M. Jackson High School P.O. Box 1409 Mill Creek, WA 98012 September 5, 2013- January 18, 2014 Mr. Andrew Sevald Science Research Instructor Mill Creek, Washington 98012 2 NAME: HOME ADDRESS: Indira Rayala 3215 179th ST SE Mill Creek, WA 98012 SCHOOL: Henry M. High School SPONSOR/TEACHER: Mr. Andrew Sevald TITLE Methanol Extract of Terminalia bellirica: A Novel Quorum Sensing Inhibitor and the Application of Terminalia bellirica as an Environmental and Cost-Effective to Plant Pathogens in Crops. Abstract: 3 Contents: Page Purpose 4 Hypothesis 5 Independent and Dependent Variables 6 Introduction 7-8 Method 9-13 Discussion of Results Tables and Figures Attachments 14-15 16 17-20 Statistical Analysis 21 Conclusion 22 Future Studies 23 Acknowledgements 24 Bibliography 25 4 Purpose The purpose of this study was to determine: 1. If the methanol extract of Teminalia bellirica will show Anti-Quorum Sensing qualities 2. If the methanol extract of Terminalia chebula Retz will inhibit Quorum Sensing in Ralstonia Solanacearum and Xanthomonas Oryza pv. oryzae 3. If the methanol extract of Terminalia Bellirica will prevent Ralstonia Solancearum from infecting a tomato plant. In order to: i. Develop an environmental and cheap antibiotic for plant pathogens in crops. ii. Develop an antibiotic that plant pathogenic bacteria will not become resistant to. 5 Hypotheses 1. The methanol/aqueous extract from Terminalia bellirica will inhibit the bio-film production in Ralstonia solanacearum and Xanthomonas oryzae pv. oryzae 2. The methanol/aqueous extract from Terminalia bellirica will show Anti-Quorum sensing ability in the disc diffusion assay. 3. The methanol/aqueous extract from Terminalia bellirica will change the integrity of AHLs molecules Variables 1. The independent variables are the methanol and aqueous extracts of Terminalia bellirica. 2. The dependent variables in this experiment are the amount of biofilm formed and the presence of blue colonies in the disc diffusion assay. 3. The controlled variable is the environment and time. 6 Introduction Annually, 500 billion dollars are lost due to bacterial diseases in crops (Oerke et al., 1994). In the United States alone, crop losses due to plant pathogens amount to $9.1 billion dollars, while worldwide, plant diseases reduce crop productivity by 12% (Food and Agriculture Organization, 1993). Current methods to treat plant disease are treating plants with chemicals antibiotics and heat. This is done to eradicate the disease, but this method is not always successful. This treatment is also hazardous to the surrounding environment, which has cause great alarm to environmentalists. A possible treatment for stopping plant disease is shutting down the pathogen’s quorum sensing. Once thought of as simple organisms, research now shows that bacteria exhibit complex methods of communications, thus making it easier to spread disease. Quorum sensing is when bacteria release signaling chemical molecules into the environment, which are called autoinducers. Quorum sensing cells produce, detect and respond to these molecules (Taga and Bassler, 2003). This is incredibly important in pathogenic bacteria since quorum sensing plays a role in launching virulence. Quorum sensing allows bacteria to chemical determine when species density becomes high enough to overcome the host cells ability to produce an effective immune response. So if quorum sensing was to be inhibited then bacteria would be “confused”. They would not be able to launch virulence and the host’s immune system will be able to kill of the pathogens (Kievit and Iglewski, 2000). While this has been heavily research in human pathogens there has been little research with plant pathogens, in comparison. One of the main autoinducers, involved in launching virulence, in gram-negative bacteria, which envelope all of the significant plant pathogens, is N-Acyl homoserine lactones (AHLS). If this autoinducer was inhibited then the pathogen would be able to launch virulence in the plant thus causing the plant no harm. This allows the plant to resist the disease and produce anti-microbial compounds to finish the pathogen off. Although, many chemical inhibitors have 7 been discovered to inhibit AHL, they raise questions on environmental safety. To avoid this issue and to discover an environmentally friendly method to prevent plant disease through quorum sensing, methanol and aqueous extracts will be derived from the ayurvedic plant Terminalia bellirica. This plant has traditionally been used to treat heart problems, but the seeds of this plant have recently been proven to be abundant with hydrolysable tannins (Sarabhai et al.). Terminalia chebula has shown anti-quorum sensing abilities 8 Method Plant Extract Preperation Dr. Dee Denver, a specialist in mutation and genome evolution, suggested that the nematodes, Caenorhabditis elegans (C.elegans) be used as a model species for producing my Model for Sustainable Fisheries. C. elegans are eukaryotes, meaning they share cellular structures, molecular structures, and control pathways with higher organisms. C. elegans are also multicellular organisms, containing 959 cells. C. elegans go through a complex developmental process that includes embryogenesis, morphogenesis, and growth to an adult (Eisenmann 2005). Hence, C. elegans can be directly related to more complex organisms, such as fish species. Another advantage to using C. elegans in producing my model is the speed of the C. elegan life cycle. C. elegans develop from egg to adult in three days and produce a few hundred offspring three days after becoming an adult. Whereas if a fish species such as Atlantic Silversides were used, it could take years to produce enough generations so as to develop a model. I obtained my stock of C.elegans by mail from Dr. Denver, assistant professor of zoology at Oregon State University. Also In preparation for my study, I purchased forty NGM agar plates seeded with the C. elegans food source, e. coli, and forty NGM agar plates not seeded with E.coli. I also purchased a Swift* M10 Series Compound Digital Microscope with a 3-in flip up LCD screen with USB output and camera that provides 5-megapixel still images. In addition to 9 the microscope, SwiftCam Imaging II software was purchased to measure and annotate images taken by the microscope’s camera. Obtaining Synchronous Populations of C. elegans Five groups with five replicates in each were used in my study. The first two groups, A: 1-5 and B: 1-5, were used in the verification phase of my study and the last three groups, C: 1-5, D: 1-5, and E: 1-5, were used in the model phase of my study. Each of the 25 populations in my entire study had to be synchronized. That is, the C. elegans in each population had to all start at the same phase in their life cycle. In doing this, a process from www.wormbook.org; Maintenance of C. elegans, was revised and used to acquire just the eggs from a stock population of C.elegans. First, I pipetted distilled water across the top of the agar of a stock dish of C. elegans. This loosened the worms and eggs stuck in the bacteria and agar. Then, I collected 3.5-mL of the distilled water from the dish into a test tube. A solution of .5mL 5 N NaOH and 1-mL household bleach (5% solution of sodium hypochlorite) was mixed and then added to the test tube with the 3.5-mL mixture of distilled water, worms and eggs. The mixture of bleach and 5 N NaOH is used to kill all everything except the eggs in the mixture. The test tube was then capped and sealed in preparation for the vortex and centrifuge. The test tube was vortexed for three seconds, every two minutes, for ten minutes. After vortexing, the test tube was placed in the centrifuge where it was centrifuged at 1144×g for 30 seconds. The test tube was then aspirated to 0.1-mL. This collection of 0.1mL was then put into another clean test tube and had distilled water added to it up to 5-mL. This second test tube was then centrifuged at the same speed for the same amount of time and also aspirated to 0.1mL. The 0.1mL of axenized eggs released after being centrifuged 10 was pipetted across an empty, unseeded with E. coli, NGM agar dish. The eggs were kept on the unseeded agar dish until the worms reached the L1 stage, which only took about 9 hrs. When C. elegans are starved of their food source, E. coli, they can not grow past the L1 stage. The unseeded agar dish of L1 C. elegans was then gridded and sliced into 25 equal 1cm x 1cm chunks of agar. One agar chunk was then transferred to each of the 25 agar dishes seeded with E. coli. The worms then dispersed from the unseeded agar chunks to the E. coli food on the seeded dishes, where they could then continue growing in their life cycle. Before any sizing or harvesting could be preformed, the C. elegans had to remain on the E. coli seeded dishes for 2.5 days to reach their adult sizes (Eisenmann 2005). Sample Sizing After the C. elegans had grown to the adult stage and before any harvesting could occur, sample sizing was done to each group of C. elegans to determine what sizes were to be harvested from each of the populations. The sample sizing consisted of photographing 20 random C. elegans from each population and then using SwiftCam Imaging II to size each of the 20 C. elegans in each population. Normal distribution graphs were made for each of the populations, these graphs were to be used in determining what sizes of C. elegans would be harvested from each population in the Verification and Model Phases. Also, with each development of a new generation, a sample sizing of the population was taken for use in testing a difference in the average size of C. elegans in from generation to generation. That is, after harvesting, did future generations of C. elegans produce smaller adult C.elegans. Developing an Innovative New Protocol For Harvesting C. elegans 11 The NGM agar dishes that the C. elegan populations grew in were seeded with lines of E. coli bacteria (strain OP50), so as to make it easier to quickly scan through each line of bacteria for the C. elegans. Before choosing to harvest a C. elegan, it quickly had a picture of it taken by the digital microscope that was hooked up by USB input to a computer where the program, SwiftCam Imaging II, could quickly size the C. elegan in the photo. If the size of the C. elegan coordinated with the criteria for harvesting in that particular population, then it could quickly be harvested from the agar dish. Verification Phase for Producing a Model for Sustainable Fisheries In the verification phase, two groups of synchronous C. elegans were used to verify that C. elegans would follow the same evolutionary patterns as the Atlantic Silversides in Conover’s study. That is, if 90 percent of the largest C. elegans in a population (group A:1-5) are harvested, then future generations will genetically differ to be smaller . Also, a control population (group B:1-5) had a random 90 percent of it’s population harvested from each generation for the duration of the experiment (Conover and Munch 2002). Model Phase for Producing a Model for Sustainable Fisheries Three different groups(C:1-5, D:1-5, E:1-5) were used in developing my Model for Sustainable Fisheries. From the normal distribution graphs made from the sample sizing, the size requirements for the harvesting from each of the three groups were calculated. Group C had all sizes of C. elegans that fell under a half standard deviation (50%-84%) on the distribution graph, harvested from all five of its replicates. Group D had all sizes of C. elegans that fell under one third a standard deviation (50%-72.66%) on the distribution graph, harvested from all five of its 12 replicates. Group E had all sizes of C. elegans that fell under one quarter (50%-67%) of a standard deviation on the distribution graph, harvested from all five of its replicates. Harvesting for Producing a Model for Sustainable Fisheries After the first harvest from Generation 1 of each of the groups, all of the C. elegan populations could only be harvested every six days. It takes three days for the remaining adults to reproduce and three days for the eggs produced by the remaining adults, to become adults themselves (Eisenmann 2005). To allow for the growth and harvest from five full generations of C. elegans, use of the new protocol for harvesting C. elegans and producing a model for sustainable fisheries is ongoing with results expected by March 16, 2010. 13 Discussion of Results Sample Sizing I arranged the sample sizings of all of my populations together into a list of 50 different sizes of C. elegans. The results found by my sample sizing were arranged into a standard distribution table and standard distribution graph. The standard distribution graph and table were used for representing all five populations of C.elegans. Swift* M10 Series Compound Digital Microscope and SwiftCam Imaging II The use of the Swift* M10 Series Compound Digital Microscope and SwiftCam Imaging II software was a success in being able to photograph and size the microorganism, C. elegans, in real time. The speed of sizing a C. elegan would easily allow for a quick determination of whether or not to harvest. Verification Phase 90 percent of the C. elegans from each of the Group A population replicates,1-5, are to be harvested. All C. elegans harvested in the 90 percent must be larger than a half standard deviation, or as large as the C. elegans in the top 34 % of the population distribution table and 14 graph. 90 percent of all C.elegans that meet the following criteria on the standard deviation graph, are currently being harvested. Model Phase The following are requirements for harvesting from each of the groups in the Model Phase: The size requirements for harvesting from group C:1-5 are, from 7553.512 um-7856.252 um (one half a standard deviation, from 50%-84%). The size requirements for harvesting from group D:1-5 are, from 7553.512 um-7755.339 um (one third a standard deviation, from 50%-72.66%). The size requirements for harvesting from group E:1-5 are, from 7553.512 um-7704.882 um (one fourth a standard deviation, from 50%-67%) The following results are being used in harvesting from the C. elegan populations, C:1-5, D:1-5, and E:1-5, for use in discovering which harvested group would not lose genetic variability for length in future generations. Harvesting based on this data collection is ongoing with results expected by March 16, 2010, so as to allow for growth and harvest from five full generations of a C. elegans. 15 Tables & Figures Figure 1: A population distribution graph based on sample sizings from each population, adding up to 50 different C. elegans. Normal Distribution value Population Distribution of Adult C. elegans Size of Adult C.elegans in um Table1: Displays the sizes that will be harvested from each of the C. elegan populations. mean st. dev. 7553.512245 605.4794962 Harvest from mean to bold numbers for each group 1/2 SD 302.7397481 For Model Phase: 7856.251993 Harvesting for 16 (50%-84%) 1/3 SD (50%-72.66%) 1/4 SD (50%-67%) 201.8264987 151.369874 Group C: 1-5 7755.338744 Harvesting for Group D: 1-5 7704.882119 Harvesting for Group E: 1-5 For Verification Phase: Group A: 1-5, will harvest 90 percent of the population of C. elegans that are larger than 7856.3 Group B: 1-5, will harvest a random 90 percent of its C. elegan population. Attachments Attachment 1: 17 Attachment 1 is a photograph of the Swift* M10 Compound Digital Microscope and the SwiftCam Imaging II software. 18 Attachment 2: Attachment 2 represents the agar dishes seeded with lines of E. coli. The lines of E. coli made it faster and easier to quickly scan down each line of E. coli for the C. elegans. 19 Attachment 3: Attachment 3 displays the extraction of the C. elegans eggs, so as to synchronize all populations of C. elegans. 20 Attachment 4: Attachment 4 includes two screen shot images of two, sized C. elegans on SwiftCam Imaging II. 21 Statistical Analysis Length (um) 8579.8 8565.5 8533.9 8521.1 8509.2 8437.1 8350.6 8266.4 8236.4 8227.9 8180.1 8124.2 8029.1 7935.3 7897.5 7865.8 7807.3 7710.4 7687.6 7653.9 7595.2 7546.4 7541.1 7522.9 7522.7 7485.3 7459.6 7341.9 7336.4 7334.5 7273.6 7245.8 7245.8 7201.6 7192.3 7188.6 7185.4 7181.6 7175.7 7136 7073.6 7006.7 6914.9 6722.9 6576.8 6535.8 6518.4 6516.1 6425.4 Normal Distribution 0.000156654 0.000163007 0.000177622 0.000183767 0.000189593 0.000227181 0.000277001 0.00032945 0.000348812 0.000354344 0.000385708 0.000422573 0.000483992 0.000540099 0.00056069 0.000576827 0.000603477 0.000637135 0.000642926 0.000649892 0.000657327 0.000658841 0.000658748 0.000658045 0.000658034 0.000654719 0.000651009 0.00061985 0.00061786 0.000617162 0.000592109 0.000579063 0.000579063 0.000556489 0.000551478 0.000549461 0.000547706 0.000545609 0.000542328 0.000519469 0.000481272 0.000438233 0.000377788 0.000257134 0.000179373 0.000160444 0.000152815 0.000151825 0.000116146 mean st. dev. 7553.512245 605.4794962 1/2 SD 302.7397481 7856.251993 1/3 SD 201.8264987 7755.338744 1/4 SD 151.369874 7704.882119 Select out of populations from mean To number indicated in bold Lengths Mean Standard Error Median Mode Standard Deviation Sample Variance Kurtosis Skewness Range Minimum Maximum Sum Count Confidence Level(95.0%) 7553.512245 86.49707088 7522.7 7245.8 605.4794962 366605.4203 -0.788340823 0.046823524 2154.4 6425.4 8579.8 370122.1 50 173.9140141 22 Conclusion In developing a method for sustainable fisheries, a quick and efficient protocol was established for the sizing and harvesting of the microorganism, C. elegans. This methodology can, and will, be employed to develop a model for sustainable fisheries. The results collected from my study are currently being used for harvesting and for discovering at which threshold (1/2 standard deviation, 1/3 standard deviation, 1/4 standard deviation) C. elegans can be harvested without producing long-term genetic changes in a population’s size (specifically length), after harvesting from five generations. Such a method could be used to provide a better understanding for how to determine what size of fish in a population should be harvested so as to not produce long term genetic changes in a harvested fish population (specifically for length). 23 Future Studies In future studies, the model phase method for harvesting could be used with a fish species such as the Atlantic Silversides, the fish species used in Conover’s study. If results from such study are significant the same could be done for all species of harvested fish so as to determine at exactly what sizes fish can be harvested so as to not genetically change a population of fish. Such genetic changes that are occurring due to harvesting are extremely difficult to reverse. In fact, the only way to reverse a genetic change in a population is to stop harvesting the population completely for about twelve full generations (Conover et al. 2009). Due to the time a full generation can take in a harvested fish species, it would not be at all possible to completely stop the harvesting of a fish species due to the worldwide need of fishing industries for food and jobs. 24 Acknowledgements Dr. Dee Denver, Assistant Professor of Zoology, Oregon State University - Served as my Qualified Scientist, and donated C. elegans Mr. Chris Reeves, Science Research Instructor - Assisted with design of my study, provided help with all of my questions, allowed me access to the biology lab, reviewed my paper Miss Lori Schlatter - Provided hours of help and insight in synchronizing C. elegan populations, also provided a tremendous amount of moral support Miss Anna Morgan - Stayed very late after school to provide help and moral support Mr. Jay Gulshen - Stayed late to help and provide a minimal amount of support Mrs. Kim Griggs, Science Research Instructor - Brought a very good vibe to the Science Research environment Mrs. Paula O’Connor, Parent - For allowing me to stay at the school lab for long hours and extremely late nights. Also for bringing me lunch when I was hungry. 25 Bibliography Allendorf , F. W., England, P. R., Luikart, G., Ritchie, P. A. & Ryman, N. 2008 Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 23, 327–337. (doi:10.1016/ j.tree.2008.02.008) Conover D O, Munch S B. Sustaining Fisheries Yields Over Evolutionary Time Scales, Science 2002; 297: 94-96. Conover D O, Munch S B, Arnott S A. Reversal of evolutionary downsizing caused by selective harvest of large fish, Proceedings of the royal society 2009; 1-6. Available from: http://www.oceanconservationscience.org/press/files/rs .... Eisenmann, D. M., Wnt signaling (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.7.1, http://www.wormbook.org. Ernande B, Dieckmann U, Heino M. Adaptive changes in harvested populations : plasticity and evolution of age and size at maturation, The Royal Society 2004; 271: 415-423. Jørgensen, C. et al. 2007 Ecology: managing evolving fish stocks. Science 318,1247–1248. (doi:10.1126/science. 1148089) Oliver R.All About: Global Fishing. In: CNN [discussion list on the Internet]. 2008 Mar. 29; [cited 2009 Oct. 29]. 3 p. Available from: http://edition.cnn.com/2008/WORLD/asiapcf/03/24/eco.aboutfishing/index.html