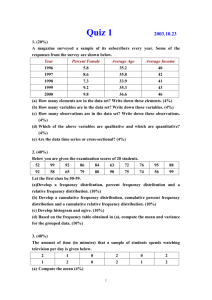
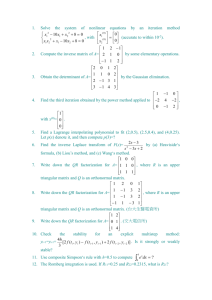
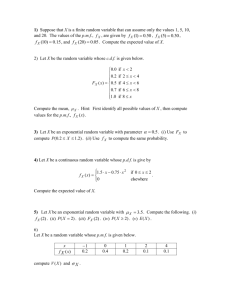
IMT4741 – Intrusion detection and prevention Exercise 3. Anomaly
advertisement
IMT4741 – Intrusion detection and prevention
Exercise 3. Anomaly detection systems
1. Suppose that IDES monitors 3 parameters of a computer system: CPU usage, memory usage
and network usage. Denote CPU usage by S1, memory usage by S2 and network usage by S3.
a) Suppose that S1, S2 and S3 are mutually correlated and that the correlation coefficients
are the following: c(S1,S2)=0.3, c(S1,S3)=0.2, c(S2,S3)=0.1. Let the values of S1, S2 and S3 be
0.2, 0.4 and 0.3, respectively, at the time instant t. Compute the IDES score of the system
at the time instant t.
b) Suppose that S1, S2 and S3 are not correlated. Compute the IDES score of the system at
the time instant t, for the same values of S1, S2 and S3 as in the case a). Compute the
NIDES score T2 for the case without correlation.
1 0.3 0.2
a) n=3. The correlation matrix C = [0.3 1 0.1]. We represent the values of the system
0.2 0.1 1
parameters in the vectorial form S = [𝑆1 , 𝑆2 , 𝑆3 ] . The IDES score 𝐼𝑆 = SC−1 ST. To
compute IS, we have to invert the correlation matrix C first. Recall from linear algebra
C−1
1 𝐶11
=
[𝐶21
det C 𝐶
31
𝐶12
𝐶22
𝐶32
𝐶13 T
𝐶23 ]
𝐶33
Now the minors Cij are:
𝑐22
𝐶11 = (−1)1+1 |𝑐
32
𝑐23
𝑐
1+2 21
|𝑐
𝑐33 |, 𝐶12 = (−1)
31
𝑐23
𝑐
1+3 21
|𝑐
𝑐33 |, 𝐶13 = (−1)
31
𝑐22
𝑐32 |
𝑐12
𝐶21 = (−1)2+1 |𝑐
32
𝑐13
𝑐
2+2 11
|𝑐
𝑐33 |, 𝐶22 = (−1)
31
𝑐13
𝑐
2+3 11
|𝑐
𝑐33 |, 𝐶23 = (−1)
31
𝑐12
𝑐32 |
𝑐12
𝐶31 = (−1)3+1 |𝑐
𝑐13
𝑐
3+2 11
(−1)
𝐶
=
|,
|
32
𝑐23
𝑐21
𝑐13
𝑐
3+3 11
(−1)
𝐶
=
|,
|
33
𝑐23
𝑐21
𝑐12
𝑐22 |
22
By means of the Kramer’s rule, we compute these minors:
𝐶11 = 𝑐22 𝑐33 − 𝑐32 𝑐23 = 1 ∙ 1 − 0.1 ∙ 0.1 = 0.99
𝐶12 = −(𝑐21 𝑐33 − 𝑐31 𝑐23 ) = −(0.3 ∙ 1 − 0.2 ∙ 0.1) = −0.28
𝐶13 = 𝑐21 𝑐32 − 𝑐31 𝑐22 = 0.3 ∙ 0.1 − 0.2 ∙ 1 = −0.17
𝐶21 = −(𝑐12 𝑐33 − 𝑐32 𝑐13 ) = −(0.3 ∙ 1 − 0.1 ∙ 0.2) = −0.28
𝐶22 = 𝑐11 𝑐33 − 𝑐31 𝑐13 = 1 ∙ 1 − 0.2 ∙ 0.2 = 0.96
𝐶23 = −(𝑐11 𝑐32 − 𝑐31 𝑐12 ) = −(1 ∙ 0.1 − 0.2 ∙ 0.3) = −0.04
𝐶31 = 𝑐12 𝑐23 − 𝑐22 𝑐13 = 0.3 ∙ 0.1 − 1 ∙ 0.2 = −0.17
𝐶32 = −(𝑐11 𝑐23 − 𝑐21 𝑐13 ) = −(1 ∙ 0.1 − 0.3 ∙ 0.2) = −0.04
𝐶33 = 𝑐11 𝑐22 − 𝑐21 𝑐12 = 1 ∙ 1 − 0.3 ∙ 0.3 = 0.91
The determinant det C is:
1 0.3 0.2 1 0.3
det 𝐂 = |0.3 1 0.1| 0.3 1 =
0.2 0.1 1 0.2 0.1
= 1 ∙ 1 ∙ 1 + 0.3 ∙ 0.1 ∙ 0.2 + 0.2 ∙ 0.3 ∙ 0.1 − 0.2 ∙ 1 ∙ 0.2 − 0.1 ∙ 0.1 ∙ 1 − 1 ∙ 0.3 ∙ 0.3 =
= 0.872
Then, the inverted correlation matrix is:
𝐂 −1
0.99 −0.28 −0.17 T
0.99 −0.28 −0.17
1
1
=
[−0.28 0.96 −0.04] =
[−0.28 0.96 −0.04] =
0.872
0.872
−0.17 −0.04 0.91
−0.17 −0.04 0.91
1.135 −0.321 −0.195
= [−0.321 1.101 −0.046]
−0.195 −0.046 1.044
We can now compute the IDES score:
𝐼𝑆 = SC−1 ST
1.135 −0.321 −0.195
𝐒𝐂 −1 = [0.2 0.4 0.3] ∙ [−0.321 1.101 −0.046] = [0.0401 0.3624 0.2363]
−0.195 −0.046 1.044
0.2
𝐼𝑆 = [0.0401 0.3624 0.2363] ∙ [0.4] = 0.22387.
0.3
1 0 0
1
b) No-correlation case means 𝐂 = 𝐈 = [0 1 0]. Then, 𝐂 −1 = 𝐈 = [0
0 0 1
0
compute IS directly:
𝐼𝑆 = SC−1 ST
1 0 0
𝐒𝐂 −1 = [0.2 0.4 0.3] ∙ [0 1 0] = [0.2 0.4 0.3]
0 0 1
0.2
𝐼𝑆 = [0.2 0.4 0.3] ∙ [0.4] = 0.29
0.3
The NIDES score T2:
1
0 0
1 0]. We can
0 1
1
𝑇 2 = 𝑛 ∑𝑛𝑗=1 𝑆𝑗2 = 3 (0.22 + 0.42 + 0.32 ) = 0.097 .
2. A clustering-based IDS that uses k-means clustering algorithm monitors 3 parameters of a
computer system: CPU usage, memory usage and network usage. The following observations
of these parameters were obtained at 10 different time intervals t1, …, t10:
𝐕1 = [0.2 0.4 0.1]
𝐕2 = [0.9 0.7 0.3]
𝐕3 = [0.9 0.7 0.2]
𝐕4 = [0.8 0.6 0.4]
𝐕5 = [0.8 0.5 0.4]
𝐕6 = [0.8 0.5 0.3]
𝐕7 = [0.2 0.2 0.2]
𝐕8 = [0.2 0.3 0.2]
𝐕9 = [0.2 0.2 0.1]
𝐕10 = [0.3 0.2 0.2]
If k=2 and the k-means algorithm performs only one iteration, determine the clusters and the
new centers of the clusters after that iteration. The initial centers are V1 and V2. Euclidean
distance is used.
The initial assignment of the given vectors:
𝑑(𝐕3 , 𝐕1 ) = √(0.9 − 0.2)2 + (0.7 − 0.4)2 + (0.2 − 0.1)2 = 0.768
𝑑(𝐕3 , 𝐕2 ) = √(0.9 − 0.9)2 + (0.7 − 0.7)2 + (0.2 − 0.3)2 = 0.1
⇒ 𝐕3 → 𝐕2
𝑑(𝐕4 , 𝐕1 ) = √(0.8 − 0.2)2 + (0.6 − 0.4)2 + (0.4 − 0.1)2 = 0.7
𝑑(𝐕4 , 𝐕2 ) = √(0.8 − 0.9)2 + (0.6 − 0.7)2 + (0.4 − 0.3)2 = 0.173
⇒ 𝐕4 → 𝐕2
𝑑(𝐕5 , 𝐕1 ) = √(0.8 − 0.2)2 + (0.5 − 0.4)2 + (0.4 − 0.1)2 = 0.678
𝑑(𝐕5 , 𝐕2 ) = √(0.8 − 0.9)2 + (0.5 − 0.7)2 + (0.4 − 0.3)2 = 0.245
⇒ 𝐕5 → 𝐕2
𝑑(𝐕6 , 𝐕1 ) = √(0.8 − 0.2)2 + (0.5 − 0.4)2 + (0.3 − 0.1)2 = 0.64
𝑑(𝐕6 , 𝐕2 ) = √(0.8 − 0.9)2 + (0.5 − 0.7)2 + (0.3 − 0.3)2 = 0.224
⇒ 𝐕6 → 𝐕2
𝑑(𝐕7 , 𝐕1 ) = √(0.2 − 0.2)2 + (0.2 − 0.4)2 + (0.2 − 0.1)2 = 0.224
𝑑(𝐕7 , 𝐕2 ) = √(0.2 − 0.9)2 + (0.2 − 0.7)2 + (0.2 − 0.3)2 = 0.866
⇒ 𝐕7 → 𝐕1
𝑑(𝐕8 , 𝐕1 ) = √(0.2 − 0.2)2 + (0.3 − 0.4)2 + (0.2 − 0.1)2 = 0.141
𝑑(𝐕8 , 𝐕2 ) = √(0.2 − 0.9)2 + (0.3 − 0.7)2 + (0.2 − 0.3)2 = 0.812
⇒ 𝐕8 → 𝐕1
𝑑(𝐕9 , 𝐕1 ) = √(0.2 − 0.2)2 + (0.2 − 0.4)2 + (0.1 − 0.1)2 = 0.2
𝑑(𝐕9 , 𝐕2 ) = √(0.2 − 0.9)2 + (0.2 − 0.7)2 + (0.1 − 0.3)2 = 0.883
⇒ 𝐕9 → 𝐕1
𝑑(𝐕10 , 𝐕1 ) = √(0.3 − 0.2)2 + (0.2 − 0.4)2 + (0.2 − 0.1)2 = 0.245
𝑑(𝐕10 , 𝐕2 ) = √(0.3 − 0.9)2 + (0.2 − 0.7)2 + (0.2 − 0.3)2 = 0.787
⇒ 𝐕10 → 𝐕1
The clusters, after the first iteration:
Cluster 1: V1, V7, V8, V9, V10
Cluster 2: V2, V3, V4, V5, V6
Calculation of the new centers:
𝐕1 = [0.2 0.4 0.1]
𝐕7 = [0.2 0.2 0.2]
𝐕8 = [0.2 0.3 0.2]
𝐕9 = [0.2 0.2 0.1]
𝐕10 = [0.3 0.2 0.2]
𝐶1′ = [0.22 0.26 0.16]
𝐕2 = [0.9 0.7 0.3]
𝐕3 = [0.9
𝐕4 = [0.8
𝐕5 = [0.8
𝐕6 = [0.8
𝐶2′ = [0.84
0.7
0.6
0.5
0.5
0.6
0.2]
0.4]
0.4]
0.3]
0.32]
3. Determine the Davies-Bouldin index of the clustering obtained in the previous example. Use
the centroid diameter for the intra-cluster distance and centroid linkage for the inter-cluster
distance. Euclidean distance is used.
𝐿
Δ(𝐶𝑖 ) + Δ(𝐶𝑗 )
1
𝐷𝐵(𝐶) = ∑ max {
}
𝑖≠𝑗
𝐿
𝛿(𝐶𝑖 , 𝐶𝑗 )
𝑖=1
Δ(𝐶𝑖 ) intra-cluster distance
δ(Ci , Cj ) inter-cluster distance
Centroid diameter:
Δ(𝐶𝑖 ) = 2
𝐬𝐶𝑖 =
∑𝐗𝑘 ∈𝐶𝑖 𝑑(𝑋𝑘 , 𝐬𝐶𝑖 )
|𝐶𝑖 |
1
∑ 𝐗𝑘
|𝐶𝑖 |
𝐗 𝑘 ∈𝐶𝑖
𝐬𝐶𝑖 is the centroid vector. 𝐗 𝑘 are the vectors of the cluster 𝐶𝑖 .
In our case = 2 , 𝐶1 = {𝐕1 , 𝐕7 , 𝐕8 , 𝐕9 , 𝐕10 } , 𝐶2 = {𝐕2 , 𝐕3 , 𝐕4 , 𝐕5 , 𝐕6 } ,
𝐬𝐶1 = 𝐶1′ = [0.22 0.26 0.16] , 𝐬𝐶2 = 𝐶2′ = [0.84 0.6 0.32].
To compute intra-cluster distances (in this case the centroid diameters), we need the
distances of all the vectors in the clusters to their corresponding centroids:
𝑑(𝐕1 , 𝐬𝐶1 ) = √(0.2 − 0.22)2 + (0.4 − 0.26)2 + (0.1 − 0.16)2 = 0.154
𝑑(𝐕7 , 𝐬𝐶1 ) = √(0.2 − 0.22)2 + (0.2 − 0.26)2 + (0.2 − 0.16)2 = 0.075
𝑑(𝐕8 , 𝐬𝐶1 ) = √(0.2 − 0.22)2 + (0.3 − 0.26)2 + (0.2 − 0.16)2 = 0.06
𝑑(𝐕9 , 𝐬𝐶1 ) = √(0.2 − 0.22)2 + (0.2 − 0.26)2 + (0.1 − 0.16)2 = 0.087
𝑑(𝐕10 , 𝐬𝐶1 ) = √(0.3 − 0.22)2 + (0.2 − 0.26)2 + (0.2 − 0.16)2 = 0.108
𝑑(𝐕2 , 𝐬𝐶2 ) = √(0.9 − 0.84)2 + (0.7 − 0.6)2 + (0.3 − 0.32)2 = 0.118
𝑑(𝐕3 , 𝐬𝐶2 ) = √(0.9 − 0.84)2 + (0.7 − 0.6)2 + (0.2 − 0.32)2 = 0.167
𝑑(𝐕4 , 𝐬𝐶2 ) = √(0.8 − 0.84)2 + (0.6 − 0.6)2 + (0.4 − 0.32)2 = 0.089
𝑑(𝐕5 , 𝐬𝐶2 ) = √(0.8 − 0.84)2 + (0.5 − 0.6)2 + (0.4 − 0.32)2 = 0.134
𝑑(𝐕6 , 𝐬𝐶2 ) = √(0.8 − 0.84)2 + (0.5 − 0.6)2 + (0.3 − 0.32)2 = 0.11
The centroid diameters are:
0.154 + 0.075 + 0.06 + 0.087 + 0.108
Δ(𝐶1 ) = 2
= 0.1936
5
0.118 + 0.167 + 0.089 + 0.134 + 0.11
Δ(𝐶2 ) = 2
= 0.2472
5
The inter-cluster distance (in this case centroid-linkage) is:
𝛿(𝐶𝑖 , 𝐶𝑗 ) = 𝑑 (𝐬𝐶𝑖 , 𝐬𝐶𝑗 )
𝛿(𝐶1 , 𝐶2 ) = √(0.22 − 0.84)2 + (0.26 − 0.6)2 + (0.16 − 0.32)2 = 0.725
Then we can compute the Davies-Bouldin index:
1 Δ(𝐶1 )+Δ(𝐶2 )
Δ(𝐶 )+Δ(𝐶1 )
+ 2
)
2
𝛿(𝐶1 ,𝐶2 )
𝛿(𝐶2 ,𝐶1 )
𝐷𝐵(𝐶) = (
= 0.608 .