Comments Genomic DNA extracted from environmental samples
advertisement

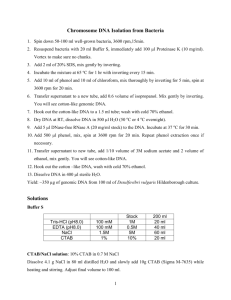
Comments Genomic DNA extracted from environmental samples, e.g. soil and sediment, contains inhibitory substances for PCR. Purifification of genomic DNA with CTAB improves PCR amplication. Basically, genomic DNA is extracted from soil with any protocols (e.g. FastDNA SPIN Kit For Soil from MP Biomedicals,USA), then purified with CTAB method. The concentration of genomic DNA is quantified with Nanodrop or Qubit assay. Finally, dilute DNA concentration to 10 ng/µL and store at -20oC for future application (Reference: Mao et al., Environ Microbiol. 2013 15:928-42). DNA purification with CTAB 1. Pre-warm CTAB (Cetyltrimethylammonium Bromide) working solution (10% CTAB/0.7 M NaCl) to 65oC in a 1.5 ml tube in the heat block. Meanwhile, proceed to step 2 and 3. 2. Transfer ~100 µL of extracted DNA into 1.5 mL tube. NOTE: If DNA volume is not 100 µL, adjust volumes of NaCl and CTAB solution accordingly. 3. Add 16.5 µL of 5 M autoclaved NaCl solution to the tube containing 100 µL DNA. 4. Add 12 µL of warm CTAB working solution. Mix thoroughly and incubate at 65 oC for 15 minutes. 5. Add 128 µL of chloroform: isoamyl alcohol (24:1). Mix carefully but thoroughly. Centrifuge at maximum speed (14,000 x g) for 5 minutes. 6. Carefully remove aqueous phase ( top layer) to a clean and labeled 1.5 ml tube (about 125 µL). Add 250 µL of cold 100% EtOH to precipitate DNA. Mix thoroughly and incubate the tube on ice or in a -20 oC for 2 hours or overnight. Centrifuge at maximum speed for 15 minutes. 7. Discard the supernatant. Add 125 µL 70% cold EtOH, flick to mix, then centrifuge at maximum speed for 5 minutes. Repeat this step one time. 8. Remove supernatant. Air dry the pellet for about 30 minutes. 9. Resuspend pellet in 100 µL ddH2O. 10. Measure the DNA concentration with Qubit Fluorometer (Invitrogen) or Nanodrop. Dilute DNA concentration to 10 ng/µL and store at -20oC.