jbi12679-sup-0001-AppendixS1-S3

Journal of Biogeography
Supporting Information:
Range-wide phylogeography of the four-toed salamander: Out of
Appalachia and into the glacial aftermath
Timothy A. Herman 1,2* and Juan L. Bouzat 1
1
Department of Biological Sciences
Bowling Green State University, Bowling Green, Ohio, USA
2
Toledo Zoo, Toledo, Ohio, USA
*TAH Current address: Indoor Ecosystems, LLC, Whitehouse, Ohio, USA
Appendix S1.
Number of samples and haplotypes detected at each collecting locality included in this study.
Appendix S2.
Observed pairwise mismatch distributions for CO1 mtDNA sequences from all samples and from individuals clades A through E.
Appendix S3.
Review of co-distributed taxa with phylogeographic patterns concordant with those resolved in Hemidactylium .
1
2
Appendix S1.
Number of samples and haplotypes detected at each collecting locality included in this study. Identification code, State, County, and Clades (including subclades) identified in the phylogenetic analysis are indicated.
Locale
Code State County
# of
Samples Haplotypes Clade (Subclade)
ALBI Alabama
ALCL Alabama
ALJA Alabama
ALLE Alabama
ALTA Alabama
ARCL Arkansas
ARGA Arkansas
ARMO Arkansas
ARPO Arkansas
FLGA Florida
FLLE Florida
FLLI Florida
FLWA Florida
GAEA Georgia
GAGR Georgia
GAHA Georgia
GAJA Georgia
GAPA Georgia
GAWA Georgia
ILJO
ILVE
Illinois
Illinois
KYAD Kentucky
KYLA Kentucky
KYME Kentucky
KYPO Kentucky
LAEF Louisiana
MDPR Maryland
MEYO Maine
MICH Michigan
MIGO Michigan
MIMO Michigan
MNIT Minnesota
MNPI Minnesota
MOBE Missouri
MODE Missouri
MOWA Missouri
MOWY Missouri
MSAD Mississippi
MSCH Mississippi
MSHI Mississippi
MSLA Mississippi
MSTI Mississippi
MSWI Mississippi
5
10
5
7
5
5
5
3
1
5
7
6
5
3
3
1
5
5
1
10
1
3
3
4
5
3
5
5
5
1
1
4
2
4
1
5
5
4
5
5
10
2
5
Bibb
Cleburne
Jackson
Lee
Tallapoosa
Cleburne
Garland
Montgomery
Polk
Gadsden
Leon
Liberty
Walton
Early
Greene
Habersham
Jasper
Paulding
Walker
Jo Daviess
Vermilion
Adair
Laurel
Menifee
Powell
East Feliciana Parish
Prince Georges/Charles
York
Chippewa
Gogebic
Montcalm
Itasca
Pine
Benton
Dent
Warren
Wayne
Adams
Chickasaw
Hinds
Lauderdale
Tishomingo
Wilkinson h1 h2 h3 h4, h5 h6, h7
B
B
D
B
B h8 h9, h10
D (Ozark)
A (Ouachita) h11 A (Ouachita) h12, h13, h14 A (Ouachita) h15, h16 h17 h18 h19
B
B
B
B h20 h21, h22 h23 h24 h25 h26 h27, h28 h27, h28
C
C
C
C
C
D
D (Midwest)
D (Midwest) h28 h28 h28, h29 h28, h30
D (Midwest)
D (Midwest)
D (Midwest)
D (Midwest), A (Teays) h31, h32 A (Southwest) h33, h34, h35 A (Northeast) h34 h28
A (Northeast)
D (Midwest) h27 D (Midwest) h28, h36, h37 D (Midwest) h27 h27, h28
D (Midwest)
D (Midwest) h38 h38 h38 h38
D (Ozark)
D (Ozark)
D (Ozark)
D (Ozark) h39 h40
A (Southwest)
A (Southwest) h41, h42, h43 A (Southwest) h44 A (Southwest) h45 h46
A (Southwest)
A (Southwest)
3
NCAV North Carolina Avery
NCBU North Carolina Buncombe
NCCH North Carolina Cherokee
NCGR North Carolina Graham
NCMA North Carolina Macon
NCOR North Carolina Orange
NCSU North Carolina Surry
NSGU Nova Scotia
NSHX Nova Scotia
Guysborough
Halifax
NYCH New York
NYSA New York
OHAD Ohio
OHAS Ohio
OHAT Ohio
OHHE Ohio
OHSC Ohio
OHWI Ohio
Chautauqua
Saratoga
Adams
Ashtabula
Athens
Henry
Scioto
Williams
OKMC Oklahoma
PAMO Pennsylvania
McCurtain
Monroe
SCOC South Carolina Oconee
SCPI South Carolina Pickens
SCYO South Carolina York
TNBL Tennessee Blount
TNHE Tennessee
TNMA Tennessee
Henry
Marion
TNMC Tennessee
TNMG Tennessee
TNMO Tennessee
TNSE Tennessee
TNSU Tennessee
TNUN Tennessee
VAAU Virginia
VACH Virginia
McMinn
Morgan
Monroe
Sevier
Sullivan
Unicoi
Augusta
Chesterfield
5
5
5
5
5
2
2
5
2
5
5
5
5
4
1
5
5
5
5
1
5
5
5
5
5
5
4
5
1
5
4
1
3 h47, h48, h49 A h50, h51 h52 h52 h53 h54 h54, h55 h34 h34
E
C
C
C
A (Northeast)
A (Northeast)
A (Northeast)
A (Northeast) h33, h34, h35 A (Northeast) h34, h54 A (Northeast) h28 h33
D (Midwest)
A (Northeast) h28 h28 h30 h27, h28
D (Midwest)
D (Midwest)
A (Teays)
D (Midwest) h13 h54 h56 h57
A (Ouachita)
A (Northeast)
C
E h58, h59 E h60, h61, h62 F h45, h63 h64, h65
A (Southwest)
D h66, h67 h68 h69, h70 h71 h72, h73 h74 h30, h33 h34
C
D
C
A
A (Teays)
A
A (Teays), A (Northeast)
A (Northeast)
VASC Virginia
WVHA West Virginia
WVKA West Virginia
Scott
Hardy
Kanawha
5
5
5 h75, h76 h30, h77 h30
A, A (Teays)
A (Teays)
A (Teays)
Acknowledgement: This study would have not been possible without the help of many individuals who collaborated with sample collection and/or assisted with identifying collection localities. We thank K.
Bekker, B. Bogaczyk, R. Bonett, J. Boundy, C. Carpenter, R. Chalmers, J. Corser, K. Enge, Z. Felix, B.
Gasdorf, J. Gilhen, I. Guenther, K. Hamed, G. Lipps, T. Majure, K. McGrath, P. Moler, J. Petranka, C.
Phillips, B. Roller, V. Schneider, J. Settles, K. Stanford, E. Timpe, and L. Williams for collaborating with sample collection. We thank R. Altig, S. Bennett, A. Braswell, A. Breisch, J. Briggler, C. Brune, C.
Camp, M. Eliott, J. Gardner, W. Gibbons, J. Gillingham, S. Graham, C. Guyer, C. Hall, S. Hall, J.
Harrison, A. Hebda, D. Hipes, J. Hohman, K. Irwin, J. Jensen, T. Johnson, R. Jones, S. Kilpatrick, T.
Mann, J. MacGregor, R. Montanucci, K. Morris, T. Pauley, R. Pfingsten, S. Roble, A. Sanders, M. Sasser,
D. Saugey, D. Stevenson, M. Sisson, J. Skeen, J. Taylor, S. Trauth, T. Walsh, and C. Wilson, for assisting with identification of collection sites.
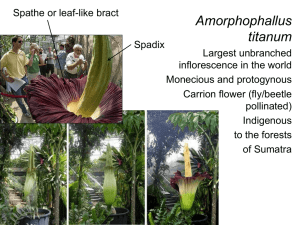
Appendix S2. Observed pairwise mismatch distributions for CO1 mtDNA sequences from all
4 samples and from individual clades A through E. Clade F was omitted due to small sample size.
Overlain on the mismatch histrogram is the expected distribution under a simulated null model of continuous expansion. High p-values indicate that the mismatch distribution fit the model of a sudden demographic expansion (All samples and Clades C and D).
Clade A - NE/SW
0.16
0.14
0.12
0.1
0.08
0.06
0.04
0.02
0
0
SSD=0.012 p=0.009
τ =13.357
2 4 6 8 10 12 14 16
# pairwise differences
18 20 22
Clade D-Midwest
SSD=0.005 p=0.273
τ =21.430
Clade B-AL/FL
0.3
0.25
0.2
0.15
0.1
0.05
SSD=0.062 p=0.001
τ =7.656
0
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
# pairwise differences
0.25
0.2
0.15
0.1
0.05
0
SSD=0.037 p=0.284
τ =8.418
0 1 2 3 4 5 6 7 8
# pairwise differences
9 10 11
Clade C-GA
0.16
0.14
0.12
0.1
0.08
0.06
0.04
0.02
0
SSD=0.017 p=0.227
τ =9.645
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
# pairwise differences
Clade E-SC
0.2
0.18
0.16
0.14
0.12
0.1
0.08
0.06
0.04
0.02
0
0 2 4
SSD=0.082 p=0.000
τ =24.492
6 8 10 12 14 16 18 20 22 24 26
# pairwise differences
Appendix S3.
Review of co-distributed taxa with phylogeographic patterns concordant with those resolved in Hemidactylium .
5
Feature
Arkansas River
Concordant Taxa
Eurycea multiplicata (R. M. Bonett and Chippindale 2004), Plethodon serratus
(Highton and Webster 1976), Plethodon caddoensis , P. fourchensis , P. kiamichi ,
P. ouachitae , P. sequoyah, P. angusticlavius , Desmognathus brimleyorum
(Conant and Collins 1998)
Mississippi embayment
Sciurus niger , S. carolinensis (Moncrief 1993), Eumeces fasciatus (Howes et al.
2006), Pantherophis obsoleta (Burbrink et al. 2000), Pantherophis guttata
(Burbrink 2002), Ambystoma maculatum (Zamudio and Savage 2003),
Etheostoma caeruleum (Ray et al. 2006), Pseudacris (Lemmon et al. 2007)
Mobile River basin
Sceloporus undulatus (Leaché and Reeder 2002), Diadophis punctatus
(Fontanella et al. 2008), Coluber constrictor (Burbrink et al. 2008), Plethodon mississippi (Highton et al. 1989), Lepomis gulosus (Bermingham and Avise 1986),
Agkistrodon piscivorus , Lampropeltis getula , Sistrurus miliarius , Trachemys scripta (Conant and Collins 1998), Narceus (Walker et al. 2009)
Apalachicola +
Chattahoochee
Rivers
Fish (Wooten and Lydeard 1990; Scribner and Avise 1993; Bermingham and
Avise 1986; Nedbal and Philipp 1994), salamanders (Donovan et al. 2000; Liu et al. 2006; Pauly et al. 2007), snakes (Burbrink et al 2000), alligator (Davis et al.
2002), turtles (Walker et al. 1995; Roman et al. 1999; Osentoski and Lamb 1995), rodents (Avise et al. 1979; Avise 1983), pine trees (Parker et al. 1997; Mylecraine et al. 2004), and pitcher plants (Godt and Hamrick 1998). See Soltis et al. (2006) for summary. Permeability of southern Apalachicola R. seen in Pseudacris crucifer (Austin et al. 2004), Pseudacris nigrita (Lemmon et al 2007),
Pantherophis obsoleta (Burbrink et al 2002), Narceus (Walker et al. 2009)
Little Tenn. +
Toxaway + Keowee
Rivers
Plethodon chlorobryonis , P. metcalfi , P. teyahalee (Highton et al. 1989; Highton and Peabody 2000), Desmognathus wrighti (Crespi et al. 2003), Fumontana deprehendor (Thomas and Hedin 2008)
French Broad River
Desmognathus wrighti (Crespi et al. 2003), Desmognathus ocoee/carolinensis
(Tilley and Mahoney 1996), Plethodon teyahalee/cylindraceus (Highton and
Peabody 2000), Aneides aeneus (Bernardo and Corser, in prep.), Fumontana deprehendor (Thomas and Hedin 2008)
Separate
Eastern/Midwestern glacial refugia
Ambystoma maculatum (Zamudio and Savage 2003), Ambystoma tigrinum
(Church et al. 2003), Eurycea bislineata (Kozak et al. 2006), Pseudacris crucifer
(Austin et al. 2002), Rana pipiens (Hoffman and Blouin 2004), Pantherophis obsoleta (Burbrink et al. 2000), Blarina brevicauda (Brant and Orti 2002),
Narceus (Walker et al. 2009)
Appendix S3 (cont.)
Feature Concordant Taxa
Southern Mississippi
River crossing
Diadophis punctatus (Fontanella et al. 2008), Sceloporus undulatus (Leaché and
Reeder 2002), quadridigitata ,
Desmognathus auriculatus
Plethodon kisatchie
, D. brimleyorum , D. conanti
(Conant and Collins 1998)
, Eurycea
6
Ozark - Cumberland
Plateau sister taxa
Cryptobranchus alleganiensis , Eurycea longicauda , Eurycea lucifuga , Plethodon dorsalis/angusticlavius, Hybopsis dissimilis , Typhlichthys subterraneus (Conant
1960), Noturus exilis (Hardy et al. 2002)
Cumberland taxa distributed across
Ohio River
Aneides aeneus , Eurycea lucifuga , Gyrinophilus porphyriticus duryi , Pseudotriton montanus diastictus (Pfingsten and Matson 2003)
Teays River associated taxa
Gyrinophilus porphyriticus duryi (R. A. Brandon 1966), Eurycea bislineata complex (Kozak et al. 2006), Plethodon kentucki , P. richmondi , Desmognathus welteri
Ancient Tennessee-
Mobile River connection
Freshwater mussels and fish (Simpson 1900; van der Schalie 1939; Wall 1968;
Swift et al. 1986; Mayden 1988; Phillips and Johnston 2004), Sternotherus
(Iverson 1977; Seidel and Lucchino 1981; Reynolds and Seidel 1983)
References
Austin, J. D., Lougheed, S.C. & Boag, P.T. (2004) Discordant temporal and geographic patterns in maternal lineages of eastern north American frogs, Rana catesbeiana (Ranidae) and
Pseudacris crucifer (Hylidae). Molecular Phylogenetics and Evolution , 32 , 799-816.
Austin, J.D., Lougheed, S.C., Neidrauer, L., Chek, A.A. & Boag, P.T. (2002) Cryptic lineages in a small frog: the post-glacial history of the spring peeper, Pseudacris crucifer (Anura:
Hylidae). Molecular Phylogenetics and Evolution , 25 , 316-329.
Avise, J.C. (1983) Mitochondrial DNA differentiation during the speciation process in
Peromyscus. Molecular Biology and Evolution , 1 , 38-56.
Avise, J.C., Giblin-Davidson, C., Laerm, J., Patton, J.C. & Lansman, R.A. (1979) Mitochondrial
DNA clones and matriarchal phylogeny within and among geographic populations of the pocket gopher, Geomys pinetis. Proceedings of the National Academy of Sciences , 76 ,
6694-6698.
Bermingham, F. & Avise, J.C. (1986) Molecular zoogeography of freshwater fishes in the
Southeastern United States. Genetics , 113 , 939-965.
Bonett, R.M. & Chippindale, P.T. (2004) Speciation, phylogeography and evolution of life history and morphology in plethodontid salamanders of the Eurycea multiplicata complex. Molecular Ecology , 13 , 1189-1203.
Brandon, R.A. (1966) Systematics of the Salamander Genus Gyrinophilus . University of Illinois
Press, Urbana, IL.
Brant, S.V. & Orti, G. (2002) Molecular phylogeny of short-tailed shrews, Blarina (Insectivora:
Soricidae). Molecular Phylogenetics and Evolution , 22 , 163-173.
Burbrink, F.T. (2002) Phylogeographic analysis of the cornsnake ( Elaphe guttata ) complex as inferred from maximum likelihood and Bayesian analyses. Molecular Phylogenetics and
Evolution , 25 , 465-476.
7
Burbrink, F.T., Fontanella, F., Alexander Pyron, R., Guiher, T.J. & Jimenez, C. (2008)
Phylogeography across a continent: The evolutionary and demographic history of the
North American racer (Serpentes: Colubridae: Coluber constrictor ). Molecular
Phylogenetics and Evolution , 47 , 274-288.
Burbrink, F.T., Lawson, R. & Slowinski, J.B. (2000) Mitochondrial DNA Phylogeography of the
Polytypic North American Rat Snake ( Elaphe obsoleta ): A Critique of the Subspecies
Concept. Evolution , 54 , 2107-2118.
Church, S.A., Kraus, J.M., Mitthell, J.C., Church, D.R. & Taylor, D.R. (2003) Evidence for multiple Pleistocene refugia in the postglacial expansion of the eastern tiger salamander,
Ambystoma tigrinum tigrinum . Evolution , 57 , 372-383.
Conant, R. (1960) The Queen Snake, Natrix septemvittata , in the Interior Highlands of Arkansas and Missouri, with comments upon similar disjunct distributions. Proceedings of the
Academy of Natural Sciences of Philadelphia , 112 , 25-40.
Conant, R. & Collins, J.T. (1998) A Field Guide to Reptiles & Amphibians: Eastern and Central
North America . Houghton Mifflin, Boston, MA.
Crespi, E.J., Rissler, L.J. & Browne, R.A. (2003) Testing Pleistocene refugia theory: phylogeographical analysis of Desmognathus wrighti , a high-elevation salamander in the southern Appalachians. Molecular Ecology , 12 , 969-984.
Davis, L.M., Glenn, T.C., Strickland, D.C., Guillette, L.J., Elsey, R.M., Rhodes, W.E., Dessauer,
H.C. & Sawyer, R.H. (2002) Microsatellite DNA analyses support an east-west phylogeographic split of American alligator populations. Journal of experimental zoology , 294 , 352-372.
Donovan, M.F., Semlitsch, R.D. & Routman, E.J. (2000) Biogeography of the southeastern
United States: a comparison of salamander phylogeographic studies. Evolution , 54 , 1449-
1456.
Godt, M.J.W. & Hamrick, J.L. (1998) Genetic divergence among infraspecific taxa of
Sarracenia purpurea . Systematic Botany , 23 , 427-438.
Hardy, M.E., Grady, J.M. & Routman, E.J. (2002) Intraspecific phylogeography of the slender madtom: the complex evolutionary history of the Central Highlands of the United States.
Molecular Ecology , 11 , 2393-2403.
Highton, R., Maha, G. & Maxson, L. (1989) Biochemical evolution in the slimy salamanders of the Plethodon glutinosus complex in the eastern United States. University of Illinois
Biological Monographs , 57 , 1-153.
8
Highton, R. & Peabody, R.B. (2000) Geographic protein variation and speciation in salamanders of the Plethodon jordani and Plethodon glutinosus complexes in the southern
Appalachian Mountains with the description of four new species. The Biology of
Plethodontid Salamanders . (ed. by R.C. Bruce, R.G. Jaeger and L.D. Houck) pp 31-93.
Kluwer Academic/Plenum Publishers, New York, NY.
Highton, R. & Webster, T.P. (1976) Geographic protein variation and divergence in populations of the salamander Plethodon cinereus . Evolution , 30 , 33-45.
Hoffman, E.A. & Blouin, M.S. (2004) Evolutionary history of the northern leopard frog: reconstruction of phylogeny, phylogeography, and historical changes in population demography from mitochondrial DNA. Evolution , 58 , 145-159.
Iverson, J.B. (1977) Geographic variation in the Musk Turtle, Sternotherus minor . Copeia , 1977 ,
502-517.
Kozak, K., Blaine, R. & Larson, A. (2006) Gene lineages and eastern North American palaeodrainage basins: phylogeography and speciation in salamanders of the Eurycea bislineata species complex. Molecular Ecology , 15 , 191-207.
Leaché, A.D. & Reeder, T.W. (2002) Molecular Systematics of the Eastern Fence Lizard
( Sceloporus undulatus ): A Comparison of Parsimony, Likelihood, and Bayesian
Approaches. Systematic Biology , 51 , 44-68.
Lemmon, E.M., Lemmon, A.R. & Cannatella, D.C. (2007) Geological and climatic forces driving speciation in the continentally distributed trilling chorus frogs (Pseudacris).
International Journal of Organic Evolution , 61 , 2086-2103.
Liu, F.G., Moler, P.E. & Miyamoto, M.M. (2006) Phylogeography of the salamander genus
Pseudobranchus in the southeastern United States. Molecular Phylogenetics and
Evolution , 39 , 149-159.
Mayden, R.L. (1988). Vicariance biogeography, parsimony, and evolution in North American freshwater fishes. Systematic zoology , 37 , 329-355.
Mylecraine, K.A., Kuser, J.E., Smouse, P.E. & Zimmermann, G.L. (2004) Geographic allozyme variation in Atlantic white-cedar, Chamaecyparis thyoides (Cupressaceae). Canadian
Journal of Forest Research , 34 , 2443-2454.
Nedbal, M.A. & Philipp, D.P. (1994) Differentiation of mitochondrial DNA in largemouth bass.
Transactions of the American Fisheries Society , 123 , 460-468.
Osentoski, M.F. & Lamb, T. (1995) Intraspecific phylogeography of the gopher tortoise,
Gopherus polyphemus : RFLP analysis of amplified mtDNA segments. Molecular
Ecology , 4 , 709-718.
Parker, K.C., Hamrick, J.L., Parker, A.J. & Stacy, E.A. (1997). Allozyme diversity in Pinus virginiana (Pinaceae): intraspecific and interspecific comparisons. American Journal of
Botany , 84 , 1372-1382.
Pauly, G.B., Piskurek, O. & Shaffer, H.B. (2007) Phylogeographic concordance in the southeastern United States: the flatwoods salamander, Ambystoma cingulatum , as a test case. Molecular Ecology , 16 , 415-29.
Pfingsten, R. & Matson, T. (2003) Ohio Salamander Atlas . Ohio Biological Survey, Columbus,
OH.
Phillips, B.W. & Johnston, C.E. (2004) Changes in the Fish Assemblage of Bear Creek
(Tennessee River Drainage) Alabama and Mississippi: 1968–2000. Southeastern
Naturalist , 3 , 205-218.
Ray, J.M., Wood, R.M. & Simons, A.M. (2006) Phylogeography and post-glacial colonization patterns of the rainbow darter, Etheostoma caeruleum (Teleostei: Percidae). Journal of
Biogeography , 33 , 1550-1558.
Reynolds, S.L. & Seidel, M.E. (1983) Morphological homogeneity in the turtle Sternotherus odoratus (Kinosternidae) throughout its range. Journal of Herpetology , 17 , 113-119.
Roman, J., Santhuff, S.D., Moler, P.E. & Bowen, B.W. (1999) Population Structure and Cryptic
9
Evolutionary Units in the Alligator Snapping Turtle. Conservation Biology , 13 , 135-142. van der Schalie, H. (1939) Additional notes on the naiades (freshwater mussels) of the lower
Tennessee River. American Midland Naturalist , 22 , 452-457.
Scribner, K.T. & Avise, J.C. (1993) Cytonuclear genetic architecture in mosquitofish populations and the possible roles of introgressive hybridization. Molecular Ecology , 2 , 139-149.
Seidel, M.E. & Lucchino, R.V. (1981) Allozymic and morphological variation among the musk turtles Sternotherus carinatus , S. depressus , and S. minor (Kinosternidae). Copeia , 1981,
119-128.
Simpson, C.T. (1900) On the evidence of the Unionidae regarding the former courses of the
Tennessee and other southern rivers. Science , 12 , 133-136.
Soltis, D., Morris, A., McLachlan, J., Manos, P. & Soltis, P. (2006) Comparative phylogeography of unglaciated eastern North America. Molecular Ecology , 15 , 4261-
4293.
Thomas, S.M. & Hedin, M. (2008) Multigenic phylogeographic divergence in the paleoendemic southern Appalachian opilionid Fumontana deprehendor Shear (Opiliones, Laniatores,
Triaenonychidae). Molecular Phylogenetics and Evolution , 46 , 645-658.
Tilley, S. & Mahoney, M. (1996) Patterns of genetic differentiation in salamanders of the
Desmognathus ochrophaeus complex (Amphibia: Plethodontidae). Herpetological
Monographs , 10 , 1-42.
Walker, D., Burke, V.J., Barak, I. & Avise, J.C. (1995) A comparison of mtDNA restriction sites vs. control region sequences in phylogeographic assessment of the musk turtle( Sternotherus minor ). Molecular Ecology , 4 , 365-373.
Walker, M.J., Stockman, A.K., Marek, P.E. & Bond, J.E. (2009) Pleistocene glacial refugia across the Appalachian Mountains and coastal plain in the millipede genus Narceus :
Evidence from population genetic, phylogeographic, and paleoclimatic data. BMC
Evolutionary Biology, 9 , 25
Wall, B.R. (1968) Studies on the fishes of the Bear Creek system of the Tennessee
River drainage . University of Alabama, Tuscaloosa, AL.
Wooten, M.C. & Lydeard, C. (1990) Allozyme variation in a natural contact zone between
Gambusia affinis and Gambusia holbrooki .
Biochemical systematics and ecology , 18 ,
169-173.
Zamudio, K.R. & Savage, W.K. (2003) Historical isolation, range expansion, and secondary contact of two highly divergent mitochondrial lineages in spotted salamanders
( Ambystoma maculatum ). Evolution , 57 , 1631-1652.