Computer Lab Document2012
advertisement

Logging into the 6th floor Computer Lab computers:
Account name: p8158. Password: p8158 (or if that doesn’t work try Carlos2177)
This document contains several modules:
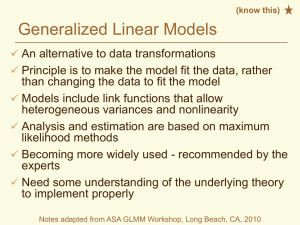
1. Using Mplus Demo Version to get and plot eigenvalues for 5 of items in the
concerns for genetic testing questionnaire that “loaded” on a single factor.
2. Using Mplus to calculate the Cronbach’s alpha for these 5 items
3. Using SAS %macro parallel to perform parallel analysis on 20 items of
concern for genetic testing questionnaire
4. Using SAS Proc Corr to calculate the Cronbach’s alpha for the 5 item
subscale
The data to be used for these analyses are available from my website
http://www.columbia.edu/~mmw2177/LVcourse/DATA/index.html
“genetictestingrawdata.xls”
1
1. Using Mplus Demo Version to get and plot eigenvalues for 5 of items in the
concerns for genetic testing questionnaire that “loaded” on a single factor.
Title: Exploratory factor analysis of genetic testing data
!In Mplus it is necessary to make each line of the code no lonter than 80 chara
!cters. There is a Column number on the lower right of the editor window.
!Exclamation points are used to COMMENT out code
Data:
file is
"C:\Users\Melanie\Documents\Teaching\LVSEM\
DataLabs\genetictestingrawdatanonames.csv";
!Data has to be simple text with either space or comma delimiting
!The dataset cannot contain the variable names. Instead
!the variable names are listed here in the program in the exact same order as
!columns in the data;
!By default in Mplus it USES the Full Information Maximum Likelihood Method
!RATHER than Listwise Deletion.
!This dataset has n=223 individuals
!If listwise deletion is used, only n=205 individuals are analyzed
!listwise = on;
variable:
names are
C1 C2 C3 C4 C5 C6 C7 C8 C9 C10 C11 C12
C13 C14 C15 C16 C17 C18 C19 C20 age race
marital educlevl insured;
usevariables are
!c1-c20;
c2 c4 c5 c6 c15;
!categorical are c2 c4 c5 c6 c15;
missing are .;
analysis: type = efa 1 2;
plot: type is plot2;
2
2. Using Mplus to calculate the Cronbach’s alpha for these 5 items
Title: Calculating Cronbach alpha
Data:
file is
"C:\Users\Melanie\Documents\Teaching\LVSEM\
DataLabs\genetictestingrawdatanonames.csv";
listwise = on;
variable:
names are
C1 C2 C3 C4 C5 C6 C7 C8 C9 C10 C11 C12
C13 C14 C15 C16 C17 C18 C19 C20 age race
marital educlevl insured;
usevariables are
c2 c4 c5 c6 c15;
missing are .;
model: f by c2@1 c4@1 c5@1 c6@1 c15@1;
!By default Mplus allows the fixed means to be different
!across the different measures (matching the two-way ANOVA mixed effect;
!If we want a one-way ANOVA result, need to fix the means to be the same
;[c4*](mu);[c5*](mu);[c6*](mu);[c15*](mu);
f*(varf);
c2*(vare);c4*(vare);c5*(vare);c6*(vare);c15*(vare);
Model constraint:
new (cronbach);
cronbach = (varf)/(varf + vare/5);
3
3. Using SAS %macro parallel to perform parallel analysis on 20 items of
concern for genetic testing questionnaire
First run the following Macro so it is loaded. Copy paste from the line that
says %macro parallel to the line that says %mend it into the editor and hit
run.
%macro parallel(data=_LAST_, var=_NUMERIC_,niter=1000, statistic=Median);
/*--------------------------------------*
| Macro Parallel |
| Parameters |
| data = dataset to be analyzed |
| (default: _LAST_) |
| var = variables to be analyzed |
| (default: _NUMERIC_) |
| niter= number of simulated datasets |
| to create (default: 1000) |
| statistic = statistic used to |
| summarized eigenvalues |
| (default: Median. Other |
| possible values: P90, |
| P95, P99) |
| Output |
| Graph of actual vs. simulated |
| eigenvalues |
*--------------------------------------*/
data _temp;
set &data;
keep &var;
run;
/* obtain number of observations and
variables in dataset */
ods output Attributes=Params;
ods listing close;
proc contents data=_temp ;
run;
ods listing;
data _NULL_;
set Params;
if Label2 eq 'Observations' then
call
symput('Nobs',Trim(Left(nValue2)));
else if Label2 eq 'Variables' then
call
symput('NVar',Trim(Left(nValue2)));
run;
/* obtain eigenvalues for actual data */
proc factor data=_temp nfact=&nvar noprint
outstat=E1(where=(_TYPE_ = 'EIGENVAL'));
var &var;
run;
data E1;
set E1;
array A1{&nvar} &var;
array A2{&nvar} X1-X&nvar;
do J = 1 to &nvar;
A2{J} = A1{J};
end;
keep X1-X&nvar;
run;
/* generate simulated datasets and obtain
eigenvalues */
%DO K = 1 %TO &niter;
data raw;
array X {&nvar} X1-X&nvar;
keep X1-X&nvar;
4
do N = 1 to &nobs;
do I = 1 to &nvar;
X{I} = rannor(-1);
end;
output;
end;
run;
proc factor data=raw nfact=&nvar noprint
outstat=E(where=(_TYPE_ =
'EIGENVAL'));
var X1-X&nvar;
proc append base=Eigen
data=E(keep=X1-X&nvar);
run;
%END;
/* summarize eigenvalues for simulated
datasets */
proc means data=Eigen noprint;
var X1-X&nvar;
output out=Simulated(keep=X1-X&nvar)
&statistic=;
proc datasets nolist;
delete Eigen;
proc transpose data=E1 out=E1;
run;
proc transpose data=Simulated out=Simulated;
run;
/* plot actual vs. simulated eigenvalues */
data plotdata;
length Type $ 9;
Position+1;
if Position eq (&nvar + 1)
then Position = 1;
set E1(IN=A)
Simulated(IN=B);
if A then Type = 'Actual';
if B then Type = 'Simulated';
rename Col1 = Eigenvalue;
run;
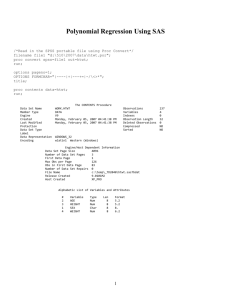
title height=1.5 "Parallel Analysis &statistic Simulated Eigenvalues";
title2 height=1 "&nvar Variables, &niter
Iterations, &nobs Observations";
proc print data = plotdata;
run;
symbol1 interpol = join value=diamond height=1 line=1 color=blue;
symbol2 interpol = join value=circle height=1 line=3 color=red
;
proc gplot data = plotdata;
plot Eigenvalue * Position = Type;
run;
quit;
%mend parallel;
Here is where you read in the data and run the parallel analysis
PROC IMPORT OUT= WORK.A DATAFILE=
"C:\Users\Melanie\Documents\Teaching\LVSEM\DataLabs\genetictestingrawdata.xls"
DBMS=EXCEL2000 REPLACE;
GETNAMES=YES; RUN;
options nocenter;
%parallel(data=a, var=c1-c20, niter=100, statistic=P95);
5
4. Using SAS Proc Corr to calculate the Cronbach’s alpha for the 5 item
subscale
PROC IMPORT OUT= WORK.A DATAFILE=
"C:\Users\Melanie\Documents\Teaching\LVSEM\DataLabs\genetictestingrawdata.xls"
DBMS=EXCEL2000 REPLACE;
GETNAMES=YES; RUN;
proc corr data = a alpha nomiss;
var c2 c4 c5 c6 c15;
run;
6