a. API
advertisement
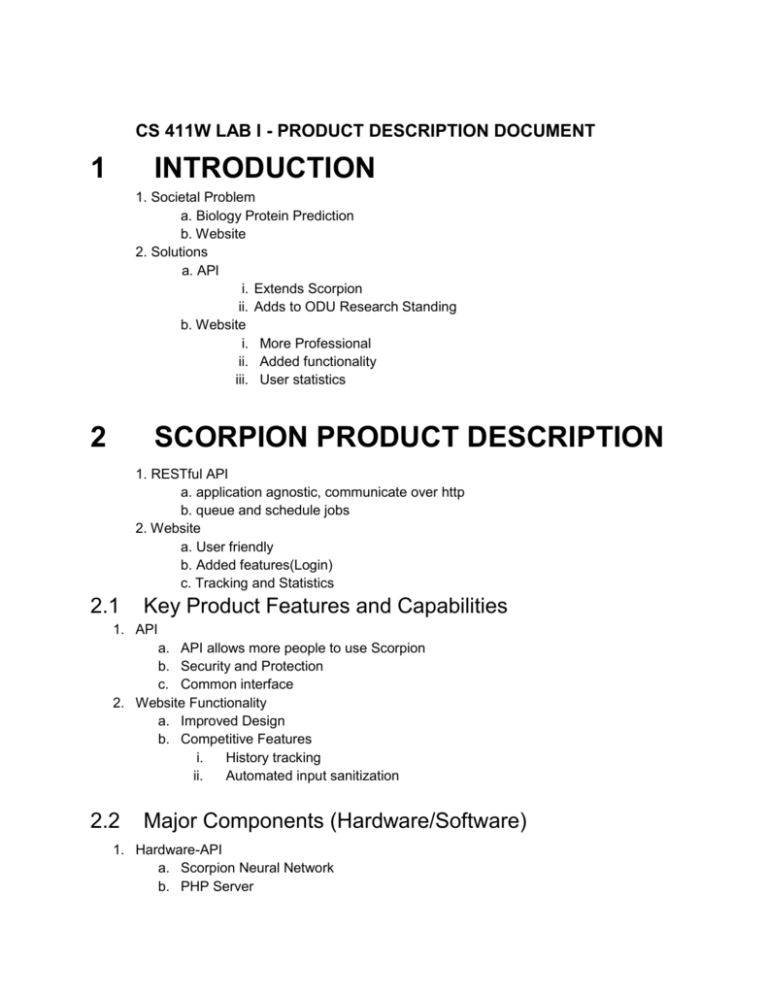
CS 411W LAB I - PRODUCT DESCRIPTION DOCUMENT 1 INTRODUCTION 1. Societal Problem a. Biology Protein Prediction b. Website 2. Solutions a. API i. Extends Scorpion ii. Adds to ODU Research Standing b. Website i. More Professional ii. Added functionality iii. User statistics 2 SCORPION PRODUCT DESCRIPTION 1. RESTful API a. application agnostic, communicate over http b. queue and schedule jobs 2. Website a. User friendly b. Added features(Login) c. Tracking and Statistics 2.1 Key Product Features and Capabilities 1. API a. API allows more people to use Scorpion b. Security and Protection c. Common interface 2. Website Functionality a. Improved Design b. Competitive Features i. History tracking ii. Automated input sanitization 2.2 Major Components (Hardware/Software) 1. Hardware-API a. Scorpion Neural Network b. PHP Server 2. 1. 2. 3. 3 c. PSI-BLAST d. PC Software/Virtual- API a. RCSB Protein Database-Provided b. Dunbrack Labs Protein Database - Provided c. RESTful API i. GET/POST ii. Input Sanitization iii. Queueing jobs Hardware - Website a. PC b. PHP Web Server c. PSI-BLAST (Third Party/Provided) d. Scorpion Neural Network Software/Virtual - Website a. Web browser + Internet connection b. Webpage design template c. Google Analytics (Third Party/Provided) d. Database i. Protein sequence & user database 1. Open ID table 2. Sequence submissions table 3. User data table 4. User sequence submissions link table Algorithms - Website a. Protein sequence submission i. Input sanitation ii. Estimated prediction time IDENTIFICATION OF CASE STUDY 1. Scorpion a. History b. Highest Accuracy 2. Target Audience a. Professor Li b. Biologist around the world (overall scientific knowledge) c. More researchers 4 SCORPION PRODUCT PROTOTYPE DESCRIPTION 1. Interface a. protein chain input - no reduced functionality i. data sanitization ii. character counting to produce estimated time of prediction 2. PROTOTYPE FUNCTIONAL GOALS and OBJECTIVES 1. Improve the interface to scorpion a. Website i. Improved website 508 compliance b. API i. platform agnostic, using an API, users can use their language and platform of choice over http. Allows a hook to SCORPION 4.1 Prototype Architecture (Hardware/Software) <How will the prototype be structured to demonstrate key features of the 410 product. Prototype MFCD provided and described.> 1. API a. API -Backend i. Dr.Li’s c3Scorpion Binary ii. Handling POST Requests 1. Security (Attacks) 2. General POST Handling -PHP 3. Returning Job ID and Information -PHP iii. Sending Data through the NN iv. Getting Job Information 1. One Job 2. All Current Jobs v. Emailing Result b. API - Frontend i. GET format of return 1. XML/JSON 2. User Friendly 2. Website a. Server-side code in PHP i. Already used in the website before solution ii. Module based for ease of addition/removal of functions b. SQL i. Standard in database profession ii. Why SQL instead of NOSQL iii. Sqlite3 chosen for ease of use iv. Separate database from training part of project c. HTML/Javascript i. Benefits - Uses client processor instead of server ii. JQuery - Standard in web development iii. Openid.net for login - A known product used by other websites 4.2 Prototype Features and Capabilities Demonstrates 1. API a. b. c. d. Submit and schedule requests over http Get Status of Jobs over on queue Get status of a single job Public Documentation for the public to use 2. Website a. Web layout in compliance with 508 web standards b. Offer client tools for users to sanitize input c. Opt in User registration system d. User tracking solution to track registered and unregistered users usage Mitigate Risk 1. backwards compatible with existing C3 Scorpion system 2. Use open source and free tools solutions to solve user registration and tracking 4.3 Prototype Development Challenges 1. Website a. 508 compliance b. New, unfamiliar webpage design c. Accuracy of estimated prediction time d. User security 2. API a. Public facing API has to be secured b. Interfacing with existing scorpion infrastructure and codebase c. Ensuring that Admin can access statistics Glossary: Amino Acids/Residues: The building blocks of proteins Training set: Set of instances from the problem domain used to train the algorithm. Data cleansing: The process of removing non-representative instances from the data set. Cross-validation Training: The process of dividing training data into k mutually exclusive subsets (folds), of roughly equal size where some subsets are used for training, validating, and testing. The process is repeated k times. PSSM: Position-Specific Scoring Matrix which includes information about evolutionary relatives of the original protein sequence PSI-BLAST: Position-Specific Iterative Basic Local Alignment Search Tool used for deriving the PSSM References: 1. "Biological Macromolecular Resource." RCSB Protein Data Bank. N.p., n.d. Web. 20 Feb. 2014. <http://www.rcsb.org/pdb/home/home.do>. 2. CS410 Blue Team. "Scorpion Protein Prediction Timed Experiment." N.p., 11 Feb. 2014. Web. <www.cs.odu.edu/~410blue/CS410ScorpionProteinPredictionTimeEx periment.xlsx>. 3. Ashraf Yaseen and Yaohang Li. “Context-based Features Enhance Protein Secondary Structure Prediction Accuracy”. 4. "Section 508." United States Department of Health and Human Services. N.p., n.d. Web. 15 Mar. 2014. <http://www.hhs.gov/web/508/index.html>. 5. "Section 508 Of The Rehabilitation Act." Section 508 Home. N.p., n.d. Web. 15 Mar. 2014. <http://www.section508.gov/Section-508-Of-The-RehabilitationAct>.