File
advertisement

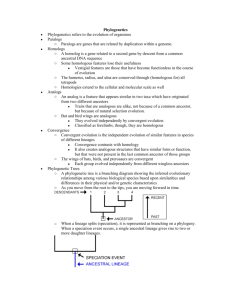
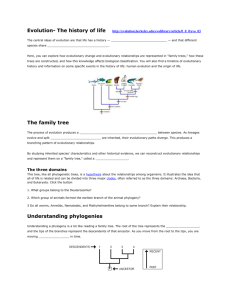
Phylogeny Systematics Taxonomy Ambiguity in everyday names Binomial Genus Specific epithet Taxon Phylogenic tree Misclassification Issue with Linnaean system PhyloCode Disadvantages Chapter 26: Phylogeny and the Tree of Life Evolutionary history of a species or a group of species A practice focused on classifying organisms and determining their evolutionary relationships; used to construct phylogenies; use data ranging from fossils to molecules & genes to infer evolutionary relationships 26.1 Phylogenies Show Evolutionary Relationships Organisms share many characteristics because of common ancestry, since an organism likely shares many of its genes, metabolic pathways, & structural proteins with its close relatives How organisms are named & classified Binomial Nomenclature Common, everyday names for organisms may refer to >1 species, and sometimes they don’t accurately reflect the kind of organism they signify Two-part format of the scientific name; came up by Carolus Linnaeus First part of the name to which the species belongs Unique for each species within the genus First letter of genus is capitalized & entire binomial is italicized Hierarchical Classification Linnaeus also grouped species into a hierarchy of increasingly inclusive categories Species that appear to be closely related are grouped into the same genus o Ex: leopard, African lion, tiger, & jaguar belong in same genus Related genera are place into the same family Related families are placed into orders Related orders are placed into classes Related classes are placed into kingdoms Related kingdoms are placed into domains Did King Phillip Come Over For Great Sex? Named taxonomic unit at any level of the hierarchy Characters useful for classifying one group of organisms may not be appropriate for other organisms; larger categories often aren’t comparable between lineages The placement of species into orders, classes, phyla, kingdoms, & domains don’t necessarily reflect evolutionary history Linking Classification and Phylogeny Branching diagram that can be used to represent evolutionary history of a group of organisms The branching pattern often matches how taxonomists have classified groups of organisms nested within more inclusive groups But sometimes, taxonomists have placed a species within a genus to which it is not most closely related. One reason for misclassification may be that over the course of evolution, a species has lost a key feature shared by its relatives Doesn’t tell us anything about the groups’ evolutionary relationships to one another Such difficulties in aligning Linnaean classification with phylogeny have led proposals that classification be based entirely on evolutionary relationships Only names groups that include a common ancestors & all of its descendants Changes the way taxa are defined, but groups would no longer have ranks attached to them, like family or class. Some commonly recognized groups would become part of other groups previously of the same rank for both classification systems Ex: because birds evolved from a group of reptiles, Aves would be considered a subgroup of Reptilia Phylogenetic tree reps a hypothesis about evolutionary relationships These relationships often are depicted as a series of dichotomies, two-way branch points Each branch point reps the divergence of 2 evolutionary lineages from a common ancestor Key points about phylogenetic trees Practical applications Ex: maize (corn) Ex: what is the species identity of food being sold as whale meat? Taxa B & C are sister taxa, groups of organisms that share an immediate common ancestor, & thus are each other’s closest relatives This tree is rooted- a branch point within the tree reps the most recent common ancestor of all taxa in the tree Basal taxon- lineage that diverges early in the history of a group & lies on a branch that originates near the common ancestor of the group (like G lies on a branch that originates near the common ancestor of the group) Lineage leading to taxa D – F includes a polytomy- a branch point from which >2 descendant groups emerge; signifies that evolutionary relationships among the taxa are not yet clear What We Can & Can’t Learn From Phylogenetic Trees 1. They’re intended to show patterns of descent, not phenotypic similarity a. Although closely related organisms often resemble one another due to common ancestry, the may not if their lineages have evolved at different rates i. Ex: even though crocodiles are more closely related to birds than lizards, they look more like lizards because morphology has changed dramatically the bird lineage 2. The sequence of branching in a tree doesn’t necessarily determine the actual (absolute) ages of the particular species a. Doesn’t show when a species (e.g. wolf) evolved more recently than another (e.g. otter) b. Shows only that the most recent common ancestor of the two species lived before the most recent common ancestor of the wolf & coyote (2). c. To indicate when wolves & otters evolved, the tree would need to include additional divergences in each evolutionary lineage & the dates of when those splits occurred d. Usually, if unspecified, the line length is not proportional to time 3. Don’t assume that a taxon on a phylogenetic tree evolved from the taxon next to it Adding Phylogenies form a phylogeny of maize based on DNA data, 2 species of wild grasses may be maize’s closest living relatives. These 2 relatives may be useful as reservoirs of beneficial alleles that can be transferred so cultivated maize by crossbreeding or genetic engineering another use of phylogenetic trees was used to investigate whether whale meat samples had been illegally harvested from whale species protected under law Homologies Ex: mammals Ex: molecular scale Similarity in ancestry Ex: Hawaiian flowers Analogy Convergent evolution Ex: mole-like animals Distinguishing between homology & analogy Ex: bats & birds Scientists bought 13 samples of whale meat from Japanese fish markets. They sequenced a specific part of the mitochondrial DNA from each sample & compared their results with the comparable DNA sequence from known whale species To identify the species of the sample, a phylogenetic tree was constructed, showing patterns of relatedness among DNA sequences rather than among taxa The phylogeny indicated that meat from humpack, fin, & Minke whaels caught in the northern hemisphere was being sold illegally in some Japanese fish markets 26.2 Phylogenies are Inferred from Morphological & Molecular Data To infer phylogeny, info about morphology, genes, & biochemistry of the organisms, which are all features that result from common ancestry, of which these features reflect evolutionary relationships Morphological and Molecular Homologies Phenotypic & genetic similarities due to shared ancestry Similarity in number & arrangement of bones in the forelimbs of mammals is due to their descent from a common ancestor Genes or other DNA sequences are homologies if they’re descended from sequences carried by a common ancestor Organisms that share very similar morphologies or similar DNA sequences are more likely to be more closely related than organisms with vastly different structures or sequences But sometimes, morphological divergence between related species can be great & their genetic divergence small These species vary dramatically in appearance throughout the islands, but their genes are very similar Molecular data can be used to estimate divergence times Sorting Homology from Analogy Similarity due to convergent evolution Similar environmental pressures & natural selection produce similar (analogous) adaptations in organisms from different evolutionary lineages 2 mole like animals are very similar in external appearance, but their internal anatomy, physiology, & reproductive systems are very dissimilar: Australian mole are marsupials (their young complete their embryonic development in a pouch on the outside of the mother’s body), while North American moles are eutherians (their young complete their embryonic development in the uterus within the mother’s body) Genetic comparisons & the fossil record show that the common ancestor of these moles lived 140 million years ago, about the time when marsupial & eutherian mammals diverged This common ancestor & most of its descendants weren’t mole-like, but analogous characteristics evolved independently in these 2 mole lineages Distinguishing between homology & analogy is critical in reconstructing phylogenies Both birds & bats have adaptations that enable flight, which implies that bats are more closely related to birds than they are to cats, which can’t fly. But a closer examination reveals that a bat’s wing is more similar to the forelimbs of cats & other mammals than to a bird’s wing Bats & birds descended from a common tetrapod ancestry that lived ~320 million years ago. This common ancestor couldn’t fly. Although the skeletal systems of bats & birds are homologous, their wings aren’t Fossil evidence documents that bat & bird wings arose independently from the Homoplasies Complexity of comparisons Ex: skulls Ex: gene level After sequencing molecules Deceptive similarities Solutions Distinguishing between analogy & homology Cladistics Forelimbs of different tetrapod ancestors A bat’s wing is analogous, not homologous to a bird’s wing Analogous structures that arose independently The complexity of the characters being compared helps us distinguish between homology & analogy as well. The more elements that are similar in the 2 complex structures, the more likely it is that they evolved from a common ancestor Skulls of adult human & adult chimpanzee both consist of many bones fused together. The compositions of the skulls match almost perfectly. It is unlikely that such complex structures that match in so many details have separate origins. The genes involved in the development of both skulls were inherited from a common ancestor If genes in 2 organisms share many portions of their nucleotide sequences, it’s likely that the genes are homologous Evaluating Molecular Homologies Aligning comparable sequences from the species being studied. If the sequences differ only at a few sites, the species are likely very closely related, while distantly related species have different bases at many sites and different lengths, because insertions & deletions accumulate over long periods of time Two sequences could appear very similar, i.e. the first base of one sequence has been deleted, shifting the sequence back one nucleotide Computer programs have been constructed that estimate the best way to align comparable DNA segments of differing lengths These molecular comparisons reveal that many differences have accumulated in the comparable genes of Australian & North America mole. The differences indicate that their lineages have diverged a lot since their common ancestor, so they’re not closely related But the high degree of gene sequence similarity among the silverswords indicates that they’re very closely related, despite their morphological differences Two sequences that resemble each other at many points along their length most likely are homologous, but in organisms that don’t appear closely related, the bases may simply be coincidentally similar to another sequence, (molecular homoplasies) Discipline that uses data from DNA & other molecules to determine evolutionary relationships 26.3 Shared Characters are Used to Construct Phylogenetic Trees Only homology reflects evolutionary history Cladistics Common ancestry is the primary criterion used to classify organisms Biologists attempt to place species into groups called clades, each of which includes an ancestral species & all of its descendants Nested Ex: cats & dogs Monophyletic Paraphyletic Polyphyletic Shared ancestral character Ex: mammals & backbones Shared derived character Ex: mammals & hair Clades are nested within larger clades The cat group (Felidae) represents a clade within a larger clade (Carnivora) that also includes the dog group (Canine) A toxon is equivalent to a clade only if it is monophyletic- consists of an ancestra species and all of its descendants Consists of ancestra species and some, but not all of its descendants Includes taxa with different ancestors Shared Ancestral & Shared Derived Characters Originated in an ancestor of the taxon All mammals have backbones, but a backbone doesn’t distinguish mammals from other vertebrates (since they all have backbones). An evolutionary novelty unique to a clade Hair is a character shared by all mammals, but not found in their ancestors. Whether a particular character is considered ancestra or derived Inferring Phylogenies Using Derived Characters It is possible to determine the clade in which each shared derived character first appeared and to use that info to infer evolutionary relationships A species or group of species from an evolutionary lineage that’s known to have diverged before the lineage that includes the species we are studying (the ingroup) A suitable outgroup can be based on evidence from morphology, paleontology, embryonic, development, & gene sequences By comparing members of the ingroup with each other & with the outgroup, we can determine which characters were derived at the various branch points of vertebrate evolution Based on which characteristics are present in the members, we can identify the branch point in the clade Proportional branch lengths Phylogenetic trees with Proportional Branch Lengths So far, the lengths of the branches do not indicate the degree of evolutionary change in each lineage. The chronology represented by the branching pattern is relative rather than absolute The branch length reflects the number of changes that have taken place in a particular DNA sequence in that lineage. Branch length reflects the time since certain members diverged from a common ancestor Equal spans of time All different lineages that descend from a common ancestor have survived for the same number of years These equal spans of chronological time can be represented in a phylogenetic tree because whose branches are proportional to time This tree draws on fossil data to place branch points in context of geological time