AP Lab New 3 BLAST Directions 2016
advertisement

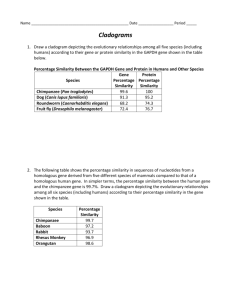
AP Lab #3 BLAST Comparing DNA Sequences Overview Watch the tutorials on the AP Bio website. Read Lab 3 in your AP lab book. We will follow that lab up to page 49. Instead of “Designing and Conducting Your Own Investigation” on page 49, we will do the Baseline Activity (page 3) from Flinn “Understanding Evolutionary Relationships” handout (Labeled “Flinn BLAST Handout” on the AP Bio website). AP Bio Lab Book Part of the lab: Before lab you will need to download 4 gene files from the site listed on page 45 of the lab. You can download those files from the site listed in your lab book, or you can download them from our AP Bio website where I have posted them. If you plan to do this activity at school you will need to bring those downloaded files with you on a USB drive, as you will not be able to download files on the school computers. You may find the Online Activities listed on pg 44 of your AP Lab Investigation Manual to be helpful. Also this may be helpful as you think of Cladistics (vs the Taxonomic Classification System you are more familiar with): Taxonomy: – Classification of living organisms into groups • Phenetic Classification System (Phylum Genus Species etc): – Groups do not necessarily reflect genetic similarity or evolutionary relatedness. Instead, groups are based on convenient, observable characteristics. • Phylogenetic Classification System (Cladograms): – Groups reflect genetic similarity and evolutionary relatedness of new (derived) characteristics. Page Directions AP Bio Lab Book 41 42 Read Answer the three questions at the bottom of the page in this cell. Which organisms have lungs? What three structures do all lizards possess? According to the cladogram, which structure — dry skin or hair — evolved first? 43 Draw the cladogram asked for at the top of the page in this cell either by hand or computer. If you will draw in by hand later, remember to leave space before you print this. Draw the cladogram for Table 1 in this cell. (either by hand or computer) 44 Answer “a” and Draw “b” a. Why is the percentage similarity in the gene always lower than the percentage similarity in the protein for each of the species? (Hint: Recall how a gene is expressed to produce a protein.) b. Draw (by hand or computer) a cladogram depicting the evolutionary relationships among all five species (including humans) according to their percentage similarity in the GAPDH gene. Record your fossil observations in this cell. Note: the last line on this page “Your task is to use BLAST to analyze these genes and determine the most likely placement of the fossil species on Figure 4” is ultimately what you are trying to figure out in this lab. 45 Form a hypothesis based on your observations. Write it here. 46 Go to the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) homepage listed and enter gene 1 (that you downloaded from the AP Bio website) following the directions given. You will come back and do the same things for genes 2-4 after you complete gene 1. 47 Read and follow directions given. 48 Answer questions 1-5 in this cell. 1. What species in the BLAST result has the most similar gene sequence to the gene of interest? 2. Where is that species located on your original cladogram? 3. How similar is that gene sequence? 4. What species has the next most similar gene sequence to the gene of interest? 5. Based on what you have learned from the sequence analysis and what you know from the structure, decide where the new fossil species belongs on the cladogram with the other organisms. If necessary, redraw the cladogram you created before. 49 Answer the four questions in “Evaluating Results” section in this cell. 1. Compare and discuss your cladogram with your classmates. Does everyone agree with the placement of the fossil specimen? If not, what is the basis of the disagreement? 2. On the main page of BLAST, click on the link “List All Genomic Databases.” How many genomes are currently available for making comparisons using BLAST? 3. How does this limitation impact the proper analysis of the gene data used in this lab? 4. What other data could be collected from the fossil specimen to help properly identify its evolutionary history? 49 “Designing and Conducting Your Investigation” you will instead Use the Flinn handout page 3 and 4, “Baseline Activity” parts A and B. Proceed below. Flinn Handout (Investigation) The basic idea of this activity is to compare the DNA sequence for the cytochrome C enzyme (which codes for a protein in the ETS) gene of multiple species, then decide the relatedness of the species based on that comparison, and depict that information in a cladogram. 1-2 3 Read Part A Steps 1-10 Go to website: http://ncbi.nlm.nih.gov then follow the directions in the Flinn lab to copy and save the sequences. Save the 13 animal sequences into your “Flinn Base Sequence Template” found on the AP Bio website BLAST lab page. 4 Part B Steps 1-9. Click to Enter the Biology Workbench 3.2 you will need to register for an account. It’s free. Use your school email. Copy the tree and paste here (or leave space and draw by hand). Q9a. Manually compare any of the four amino acid sequences you gathered from the database and count the number of differences between the human cytochrome C sequence and the four others. Does this data correlate with the phylogenetic tree? (paste the four sequences you chose to compare into this cell, then either manually or on the computer show how they compare) You will turn in this Word document (with answers and drawings filled in) and your document containing the table of all (13) the codes for all the species.