Project 3
advertisement

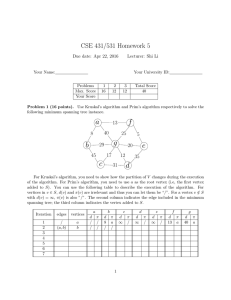
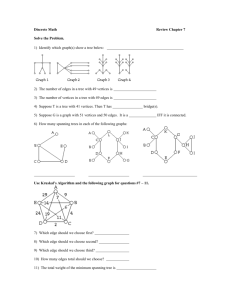
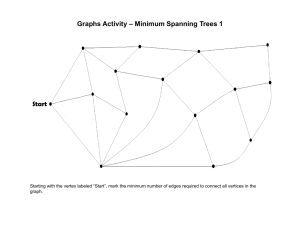
EECS 560 Project 3 Due: December 9, 2003 Description: In this project you will implement and test three different algorithms for finding the minimum spanning tree of a graph. Prim’s and Kruskal’s algorithms are described in the section 9.5 in the text, and Sollin’s algorithm is described below. In order to test the algorithms you will randomly generate graphs with a specified edge density and make sure the graphs are connected. For this part you will simply report the cost of the minimum spanning tree and the number of edges in the original graph. You must have a second version of the program that first reads the algorithm to be used for that graph (K, P or S), and then reads the edge weights of the graph. Then, find the cost of the minimum spanning tree, and print out the edges of the tree in the order they were chosen by the algorithm. Sollin’s Algorithm: Initially there are n components—one for each vertex in G Let T denote the set of edges in the spanning tree For each vertex v, add the smallest weight edge incident on v to T if it is not already in T While T contains less than n – 1 edges do For each connected component Cj of T do Find the smallest weight edge e that connects a vertex in Ci to a vertex in Cj Add e to T (if it’s not already there) Note that Sollin’s algorithm is very similar to Kruskal’s. The main difference is that it will add several edges in each pass of the while loop. Specifications: In the implementation of Kruskal's algorithm a minheap should be used to hold the edge weights. To determine if the insertion of an edge will create a cycle, use the basic data structure for the union find algorithm discussed in Section 8.3 of the text. For Prim's algorithm store the edges in adjacency lists. Use the table structure described in Section 9.5.1 to keep track of whether a vertex has already been placed in a tree and the shortest edges incident on each vertex not in the tree. You are to generate test graphs with edge densities of 20%, 50% and 100%, with 10, 20 and 100 vertices. An edge’s weight is an integer in the range 1-100. You must test a minimum of five graphs for each combination of d (edge density) and n (number of vertices). After generating a graph, perform a depth first search on it, starting from vertex 1 to see if it is connected. (Obviously, this is not necessary if the edge density is 100%.) If the DFS stops before all vertices have been listed, insert an edge between the last vertex added to the depth first search tree and the lowest numbered vertex not already in the DFS tree. Randomly generate the weight for that edge. Continue in this way until all vertices have been visited. Random numbers must be generated to determine, for each possible edge in a graph, whether the edge is present and if so, to assign it a weight. Let r be a random number in the range 0 r 1. If d = 20% and r 0.2 then the edge being considered is in the graph, so generate a second random number in the proper range to assign a weight to that edge. Similarly, if d = 50%, an edge is present if r 0.5. For d = 100%, generate a weight for each possible edge. Here are some things to watch out for: make sure you don't try to generate the same edge twice e.g. when you determine whether edge (2, 3) is in the graph do not test if edge (3, 2) should be in the graph. Be sure each algorithm uses the same data. In order to test that your algorithms are working correctly, there is a data file on the web page that has the same format the test file will have. It contains (three copies) of a complete graph (d = 100%). Run the spanning tree algorithms in the order specified in the data set. Output the cost of the minimal spanning tree and the edges of the spanning tree in the order they are chosen by the algorithm. Since this produces results different from the main project, make a second copy of the code for your spanning tree algorithms, cutting out the random graph generation, and depth first search and putting in the print statements needed to produce the output. The data file has the following format: K (or P or S) //The algorithm to use n // The number of vertices in the graph Edge weights for the edges between vertex 1 and all others in the graph. Edge weights for edges (2, 3), (2, 4), … (2, n) Edge weights for edges (3, 4), (3, 5), … (3, n) . . . A single integer--the weight of edge (n – 1, n) Y or N // Yes there’s another graph or No, you’re done. Data for another algorithm to use and graph follows if Yes Notes: There is no limit on the maximum weight of an edge. A negative value indicates that an edge is not present. “Throw away” these edges and do not include them in the data structure that holds the information for your algorithms. To turn in: 1. A copy of your well-documented code. 2. Tables that display the result of the random tests you did. (There should be 9 tables—one for each combination of d and n.) Be sure to report the information requested above for each test that you run. Do not average over the number of tests. 3. A written report that includes: a. an explanation of the data structures and how they were used for Sollin’s algorithm. b. a discussion of which of these algorithms could be used to find a minimum spanning tree for a directed graph and the changes that would be needed to handle that situation. c. a discussion of what changes would have to be made if the only data structure you were allowed to use is a two-dimensional adjacency matrix. For example, how would you determine the next edge to use? Which algorithm would be the most efficient under this condition? What is the complexity of each algorithm under this condition? d. answers to the following questions about Sollin's algorithm: 1) At least how many edges are added in the first step? 2) What is the maximum number of iterations needed? 3) What is the algorithm’s complexity? All projects and reports must be turned in by class time on the due date. No late projects will be accepted.