Biophysics Lab: Brownian Motion & Intracellular Transport
advertisement

Undergraduate Lab 111
Biophysics Experiment Matlab workbook
Brownian motion and Intracellular Transport
UC Berkeley
v1.0
This document is a live MATLAB notebook. You probably noticed that when this
document was opened, the MATLAB program was started. This is so that commands entered in
the notebook can be sent to MATLAB and evaluated. To run a command (highlighted in
green), place the cursor in it, and press Ctrl-Enter. To create your own notebook, select New
M-book from the File menu.
1. Brownian motion – a brief history
If you've ever looked through a microscope, you're probably noticed that every small
particle that you see continuously wiggles. Robert Brown, a British botanist, was not the first
person to observe these motions, but perhaps the first person to recognize the significance of this
observation [1]. Experiments quickly established the basic features of these movements. Among
other things, the magnitude of the fluctuations depended on the size of the particle, and there was
no difference between "live" objects, such as plant pollen, and things such as rock dust.
Apparently, finely crushed pieces of an Egyptian mummy also displayed these fluctuations. Says
Brown:
"[the movements] arose neither from currents in the fluid, nor from its gradual evaporation, but
belonged to the particle itself"
This effect may have remained a curiosity, had it not been for A. Einstein [2] and M.
Smoluchowski. They realized that these particle movements made perfect sense in the context of
the then developing kinetic theory of fluids. If matter was indeed composed of atoms and atoms
frequently collided with other atoms, they reasoned, then occasionally relatively large objects
such as pollen grains would exhibit random movements. This last sentence contains the
ingredients for several Nobel prizes! Indeed, Einstein's interpretation of Brownian motion as the
outcome of continuous bombardment by atoms immediately suggested a direct test of the atomic
theory of matter. J. Perrin received the 1926 Nobel Prize for validating Einstein's predictions [3],
and thus confirming the atomic theory of matter.
Since then, the field has exploded, and a through understanding of Brownian motion is
essential for everything from polymer physics to biophysics, aerodynamics, and statistical
mechanics. One of the aims of this lab is to directly reproduce the experiments of J. Perrin that
lead to his Nobel Prize. A translation of the key work is in your folder. Have a look – he used
latex spheres, and we will use polystyrene spheres, but otherwise the experiments will be
identical. The next few sections will develop the theory, as well as the Matlab code needed to
process your data.
More general info:
http://en.wikipedia.org/wiki/Brownian_motion
A great primer on particle tracking, data analysis, and diffusion:
http://www.physics.nyu.edu/grierlab/methods/methods.html
Intracellular transport
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?db=Books&rid=mcb.section.5452
Recommended reading:
Don S. Lemons, "An introduction to Stochastic Processes in Physics." The Johns Hopkins
University Press, 2002 (Intro Level)
Hans L. Pécseli, "Fluctuations in Physical Systems." Cambridge University Press, 2000
(Advanced Level)
Can we buy the above two books for the students?
Original References:
1. Brown, Robert, "A brief account of microscopical observations made in the months of June,
July and August, 1827, on the particles contained in the pollen of plants; and on the general
existence of active molecules in organic and inorganic bodies." Phil. Mag. 4, 161-173, (1828).
2. Einstein ref here
3. Perrin ref here
4. Langevin, Paul, "Sur la théorie du mouvement brownien." Comptes rendus Académie des
Sciences (Paris) 146, 530-533 (1908). Translation: AJP 65, 1079-1081 (1997).
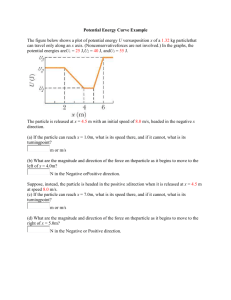
2. Simulating 1D and 2D particle trajectories
Brownian motion has been shown experimentally to consist of random displacements
whose magnitudes are normally distributed [2]. Let's simulate this process. We'll sample the
randn generator N times. The randn function samples a normal distribution with mean 0 and
standard deviation 1.
N
= 1000;
steps = randn(1,N);
plot(steps);
3
2
1
0
-1
-2
-3
-4
0
100
200
300
400
500
600
700
800
900
1000
So much for some random displacements: their sum represents a particle trajectory in 1
dimension.
position = zeros(N);
for t=2:N
position(t) = position(t-1) + randn(1, 1);
end;
plot(position);
ylabel('position');
xlabel('time steps');
title('position versus time in 1D');
position versus time in 1D
20
10
position
0
-10
-20
-30
-40
-50
0
100
200
300
400
500 600
time steps
700
800
900
1000
In two dimensions, we proceed similarly. Since all directions are (assumed to be) equivalent, all
we need to do is sample the randn generator twice per timestep.
position = zeros(2, N);
box_size = 200;
position(:,1) = (box_size * rand(1, 2)) - (box_size/2);
for t=2:N
step = [randn(1, 1), randn(1,1)];
position(:,t) = position(:,t-1) + step';
t = t+1;
end;
plot(position(1,:),position(2,:));
ylabel('position x');
xlabel('position y');
title('position versus time in 2D');
position versus time in 2D
110
100
90
position x
80
70
60
50
40
30
65
70
75
80
85
position y
90
95
100
105
3. A more realistic particle
What about a real particle? The early experiments quickly showed that the particle's diameter
and the viscosity of the embedding liquid influence the distance the particle travels. Einstein [2]
derived the precise relationship: the mean squared displacement r(t ) r(t )
2
of the particle
is proportional to the time t:
r(t ) r(t )
2
2dDt
[1]
where D is the diffusion coefficient and d is the number of dimensions. The diffusion coefficient
D, in turn, is given by
D = kBT / 6r
[length2 time-1]
[2]
where T is the temperature, kB is Boltzmann's constant, is the viscosity, and r is the particle
radius. Let's plug is some numbers. For a small particle in water, the diffusion coefficient is
about 0.2 m2/s.
r
eta
kBT
= 0.5e-6; % radius in microns
= 1.0e-3; % viscosity of water in SI units
= 1.38e-23 * 300;
dc = kBT / (6 * pi * eta * r) * 1e12
dc =
0.4393
Thus, on average, a micron-sized object in 3D travels about 2 microns in 1 second.
sqrt(2 * 3 * dc)
ans =
1.6235
There are two features of Eq. 1 worth noting – the displacement increases as the square
root of the time, and it only makes sense to speak about the average displacement, defined either
as an ensemble average (the average is taken over many particles) or as a time average (the
average is taken over many intervals of one particle trajectory1). To conclude this section, we
will generate some particle trajectories, and compute the simplest possible averages (the mean
displacement r(t ) r(t ) and the mean squared displacement r(t ) r(t )
2
). Consider
50 diffusing particles, all starting out at x = 0 at t = 0.
particles = 50;
x
= zeros(particles, N);
for t=2:N
x(:,t) = x(:,t-1) + randn(particles, 1);
end;
clf;
hold on;
for n=1:particles
plot(x(n,:));
end;
hold off;
ylabel('x(t)');
xlabel('timestep');
title('Position vs. time');
Position vs. time
100
80
60
40
x(t)
20
0
-20
-40
-60
-80
0
100
200
300
400
500 600
timestep
700
800
900
1000
What is the mean displacement? Since for each particle the probability of moving in any
particular direction is equal, the mean displacement over all particles should be zero. This will
work only if you are lucky – the number of particles we chose is still pretty small – a few
thousand would certainly be better.
mean_d = zeros(1, N);
for t=2:N
mean_d(t) = sum(x(:,t))/particles;
1
Here we're assuming that the system is ergodic.
end;
plot(mean_d);
ylabel('<d>');
xlabel('timestep');
title('<Displacement> vs. time');
<Displacement> vs. time
14
12
10
<d>
8
6
4
2
0
-2
0
100
200
300
400
500 600
timestep
700
800
900
1000
What about the mean of the square of the displacement? For every particle and time t, we need to
determine the total displacement d, (r(t) – r(0)), and then square that number, and average over
all particles. Was Einstein [2] right?
d_squared = zeros(particles, N);
mean_d_squared = zeros(N);
for t=2:N
d_squared(:,t) = x(:,t).^2;
end;
for t=2:N
mean_d_squared(t) = sum(d_squared(:,t))/particles;
end;
plot(mean_d_squared);
hold on;
plot((1:1000));
hold off;
ylabel('<d^2>');
xlabel('timestep');
title('<Displacement^2> vs. time');
2
<Displacement > vs. time
1000
800
<d2>
600
400
200
0
-200
0
100
200
300
400
500 600
timestep
700
800
900
1000
4. Estimating D from your particle trajectories
Conceptually, the simplest thing to do is to sum over many particle trajectories to obtain a
mean squared displacement, and to fit that directly to Eq. 1. Another method is described in
Grier's particle tracking primer, their Eq. 16, and involves constructing a histogram of particle
displacements for some fixed time interval, and then fitting this histogram to the expected
Gaussian distribution. Of course, since you are using the same input data for both methods, your
two estimates of D should be comparable.
Q: What values of D do you measure in your experiments? You will definitely want to vary the
particle diameter and the viscosity.
5. Auto/cross correlations
The starting point for our simulations was the assumption that we are in the overdamped
limit, or the inertia-less regime. The benefit of this assumption is that we can generate
trajectories very easily, by repeatedly querying a random number generator, and summing those
displacements to find the overall trajectory. Of course, just because it's convenient doesn't mean
it's true: do your particle trajectories display any correlations? Our simulated trajectories should
not, by construction.
How do we quantify possible correlations in a real or simulated particle trajectory? The
concept of auto-correlation was invented to quantify the degree to which a particle's trajectory in
the i'th time interval depends on some preceding time interval. If you are familiar with Markov
chains then these concepts will be second nature. The autocorrelation is defined as:
N
corr (k )
(x
i 1
i
x ) * ( xi k x )
.
N
(x
i 1
i
x)
2
Unfortunately there are a very large number of different ways of implementing this function,
depending on how rollover is handled and so forth. But mathematically, the expression is simple
– leaving normalization aside, we have:
N
corr (k ) xi * xi k
i 1
which makes sense – the product of two identical numbers (corresponding to two perfectly
correlated positions) will evaluate to a maximum. Subtracting the mean value, and dividing by N
* variance, yields the correlation coefficient, a number that ranges from 1 (perfectly correlated)
to 0 (uncorrelated). The concept of cross-correlation is an immediate extension of the concept of
autocorrelation. Here's a more practical overview.
% a correlated trajectory
x
= zeros(1,N);
c
= 0.80; % degree of correlation – 0 to 1
step = randn(1,N);
x(1) = 0;
for t=2:N
x(t) = (c * x(t-1)) + ((1-c)*step(t));
end;
plot(x);
ylabel('Displacement');
xlabel('time');
title('Displacement vs. time');
Displacement vs. time
1
0.8
0.6
Displacement
0.4
0.2
0
-0.2
-0.4
-0.6
-0.8
-1
0
100
200
300
400
500
time
600
700
800
900
1000
Applying the autocorrelation function to the trajectory, we find a correlation that decays rapidly:
intervals more than ~15 timesteps apart are uncorrelated.
K = 50;
ac = zeros(1,K);
% maximum lag
mean = sum( x ) / N;
nc
= sum( (x - mean).^2 ); % normalisation
for k=1:K+1,
ac(k) = sum( (x(1:N-k+1)-mean) .* (x(k:N)-mean) ) / nc;
end;
plot(ac);
ylabel('correlation coefficient');
xlabel('lag');
title('Autocorrelation function');
Autocorrelation function
1.2
1
correlation coefficient
0.8
0.6
0.4
0.2
0
-0.2
0
10
20
30
lag
40
50
60
6. Deviations from the Einstein model of diffusion.
In an ideal world, your experimental particle trajectories should not display any auto- orcross-correlations. In the real world, however, there are many different potential sources for such
correlations.
General questions:
1. What is a Newtonian fluid?
2. Are we really in the "inertia-less" regime? Why or why not?
3. Can the exponential term in Langevin's general solution be neglected in your
experiments? Why or why not?
4. Imagine a particle diffusing not in an isotropic material, but in a mesh or network of
some sort. Would this Einstein/Langevin description of the diffusion process still be
appropriate?
5. What would happen if particles were able to interact through some potential? Here's a
starting point.
6. What is meant by viscous coupling? Is this something you need to take into account?
Q: Are your particle trajectories auto-correlated? Over what timescales? What might lead to
autocorrelation?
Q: Are your particle trajectories cross-correlated? Over what timescales? What physical
mechanisms might lead to cross-correlation?
7. Biological transport processes: intracellular transport.
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?db=Books&rid=mcb.section.5452