Word Document
advertisement

F73DA2 INTRODUCTORY DATA ANALYSIS
Multiple regression example
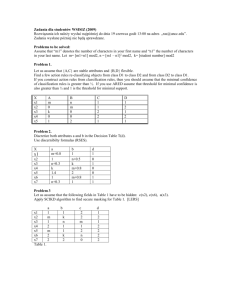
Model: Yi =xixii or E[Yi|xi] = 1x1i2x2i ; ~ N(0,2)
Data: n = 7
y
3.5
3.2
3.0
2.9
4.0
2.5
2.3
x1
3.1
3.4
3.0
3.2
3.9
2.8
2.2
x2
30
25
20
30
40
25
30
R Data are in data frame called illus2, with variables y, x1, and x2
> pairs(illus2) gives us a “matrix plot” of scatterplots for pairs of variables
3.5
2.5 3.0 3.5
3.5
2.5
y
20
x2
30
40
2.5
x1
2.5 3.0 3.5 4.0
20 25 30 35 40
.
35
3.2
3.0
Y 2.9
4.0
2.5
2.3
XX
1
1
1
1
X 1
1
1
1
31
.
3.4
3.0
3.2
39
.
2.8
2.2
30
25
20
30
40
25
30
7.0
216
.
200.0
X X 216
.
68.3 626.0
200.0 626.0 5950.0
6.68310 152925
.
0.06375
152925
.
0.76002 0.02856
0.06375 0.02856 0.00532
X X
1
2140
.
X Y 67.67
62350
.
0.213819
X Y 0.898434
0.017453
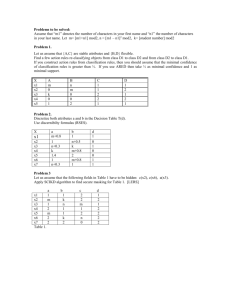
Fitted model
yˆ 0.2138 0.8984x1 0.0175x 2
Estimate of error variance 2
Y Y 67.44
X Y 671031
.
2
1
67.44 671031
0.08422
.
4
Standard errors/covariances/distributions of estimators
0.56285 0.12879 0.00537
Estimate of Cov( ˆ ) ˆ 2 ( X X ) 1 0.12879 0.06401 0.00241
0.00537 0.00241 0.00045
So:
e.s.e.( ˆ0 ) 0.750
ˆ 0 ~ N ( 0 ,0.56285)
e.s.e.( ˆ1 ) 0.253
ˆ1 ~ N ( 1 ,0.06401)
e.s.e.( ˆ2 ) 0.021
ˆ 2 ~ N ( 2 ,0.00045)
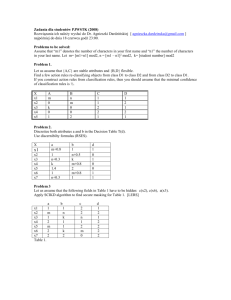
Testing the 's
Testing H: 0 = 0 we have t = -0.21382/0.56285 = - 0.29 on 4 d.f.
Similarly for 1 : t = 3.55 and for 2 : t = 0.82
Now P(| t4 | > 2.776) = 0.05 so ̂ 1 is “significantly different from 0".
We conclude that 1 0, which suggests that x1 may be a useful predictor of the response.
95% CIs for parameters
0 : -0.2138 (2.776 0.7502)
1 : 0.8984 (2.776 0.2530)
2 : 0.01745 (2.776 0.0212)
i.e. -0.2138 2.0826
i.e. 0.8984 0.7023
i.e. 0.01745 0.0589
i.e. (-2.30 , 1.87)
i.e. (0.196 , 1.60)
i.e. (-0.041 , 0.076)
CI for 2 (based on the 42 distribution of 4 2 / 2 ) is
{(4 0.08422)/11.14 , (4 0.08422)/0.4844} i.e. (0.030 , 0.695)
R
> mod2 = lm(y~x1+x2)
> summary(mod2)
Call:
lm(formula = y ~ x1 + x2)
Residuals:
1
2
0.40509 -0.07718
3
4
0.16946 -0.28475
5
6
0.01181 -0.23812
7
0.01368
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.21382
0.75024 -0.285
0.7898
x1
0.89843
0.25300
3.551
0.0238 *
x2
0.01745
0.02116
0.825
0.4558
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 0.2902 on 4 degrees of freedom
Multiple R-Squared: 0.833,
Adjusted R-squared: 0.7495
F-statistic: 9.975 on 2 and 4 DF, p-value: 0.02789
> summary.aov(mod2)
Df Sum Sq Mean Sq F value Pr(>F)
x1
1 1.62296 1.62296 19.2704 0.01178 *
x2
1 0.05730 0.05730 0.6804 0.45580
Residuals
4 0.33688 0.08422
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Fitted values v responses
3.0
2.5
mod2$fit
3.5
4.0
> plot(fitted.values(mod2)~y,pch=8)
2.5
3.0
3.5
y
4.0
Residuals v x and v fitted values
0.3
0.1
mod2$res
-0.3
-0.1
0.1
-0.3
-0.1
mod2$res
0.3
> plot(residuals(mod2)~x1,pch=8)
> plot(residuals(mod2)~x2,pch=8)
> plot(residuals(mod2)~fitted.values(mod2),pch=8)
2.5
3.0
3.5
20
25
35
x2
0.1
-0.3
-0.1
mod2$res
0.3
x1
30
2.5
3.0
3.5
4.0
mod2$fit
Using the "hat" matrix H = X(X′X)-1X′
H =
0.15270
0.10778
0.08383
0.14970
0.20060
0.12575
0.17964
0.10778
0.34984
0.35329
0.14187
0.10088
0.14533
-0.19899
0.08383
0.35329
0.49701
0.10180
-0.20359
0.24551
-0.07784
0.14970
0.14187
0.10180
0.15431
0.22985
0.11423
0.10825
3.09491
3.27718
2.83054
and then fitted values Yˆ HY 3.18475
3.98819
2.73812
2.28632
0.20060
0.10088
-0.20359
0.22985
0.80954
-0.07462
-0.06264
0.12575
0.14533
0.24551
0.11423
-0.07462
0.21442
0.22939
0.17964
-0.19899
-0.07784
0.10825
-0.06264
0.22939
0.82220
40
Interpretation of fitted models - warning
The interpretation of fitted models involving more than one explanatory variable has to be
approached with care.
We must recognise that when we examine the importance/effect of an explanatory variable, we
are examining its importance/effect in the presence of any other variables already in the model.
The coefficients/parameters and associated standard errors and P-values do not tell the whole
story and can sometimes be misleading
We need to weigh up all the evidence – all the plots and summary statistics, including the
contributions to the total sum of squares and the overall coefficient of determination.
Returning to the data above, let us examine the fits of models involving only one explanatory
variable, x1 or x2.
First x1 on its own
> mod10 = lm(y ~ x1)
> summary(mod10)
Call:
lm(formula = y ~ x1)
Residuals:
1
2
0.42868 -0.16898
3
4
0.02790 -0.27054
5
6
0.13492 -0.27366
7
0.12166
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.004506
0.683082 -0.007 0.99499
x1
0.992201
0.218681
4.537 0.00618 **
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 0.2808 on 5 degrees of freedom
Multiple R-Squared: 0.8046,
Adjusted R-squared: 0.7655
F-statistic: 20.59 on 1 and 5 DF, p-value: 0.006185
> summary.aov(mod10)
Df Sum Sq Mean Sq F value
Pr(>F)
x1
1 1.62296 1.62296 20.586 0.006185 **
Residuals
5 0.39419 0.07884
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
The total variation in the responses is Syy = 2.0171; variable x1 explains 1.6230 of this total
(80.5%) and the coefficient associated with it (0.9922) is highly significant (significantly different
from 0) – it has a very small P-value (0.006, which is < 1%). The variation explained by x1
(1.6230) is the same as in model 2 above, which was the fit of y on x1 and x2 (in that order).
Returning to the fit in model 2, we see that, in the presence of x1, x2 explains only a further
0.0573 of the total variation. Together the two variables explain 83.3% of the total variation (not
much more than the 80.5% explained by x1 alone). In the presence of x1, we gain little by
including x2.
Now x2 on its own
> mod11 = lm(y~x2)
> summary(mod11)
Call:
lm(formula = y ~ x2)
Residuals:
1
2
0.3697 0.3258
3
4
0.3818 -0.2303
5
6
7
0.3576 -0.3742 -0.8303
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.59394
1.00446
1.587
0.173
x2
0.05121
0.03445
1.486
0.197
Residual standard error: 0.5289 on 5 degrees of freedom
Multiple R-Squared: 0.3065,
Adjusted R-squared: 0.1678
F-statistic: 2.21 on 1 and 5 DF, p-value: 0.1973
> summary.aov(mod11)
Df Sum Sq Mean Sq F value Pr(>F)
x2
1 0.61820 0.61820 2.2095 0.1973
Residuals
5 1.39894 0.27979
The total variation in the responses is Syy = 2.0171 ; variable x2 explains only 0.6182 of this total
(30.6%) and the coefficient associated with it (0.0512) is not significant (not significantly
different from 0) – it has a sizeable P-value (0.197).
Back to fitting both variables: what happens is we fit x2 first, then x1?
> mod12 = lm(y~x2+x1)
> summary(mod12)
Call:
lm(formula = y ~ x2 + x1)
Residuals:
1
2
0.40509 -0.07718
3
4
0.16946 -0.28475
5
6
0.01181 -0.23812
7
0.01368
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.21382
0.75024 -0.285
0.7898
x2
0.01745
0.02116
0.825
0.4558
x1
0.89843
0.25300
3.551
0.0238 *
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
Residual standard error: 0.2902 on 4 degrees of freedom
Multiple R-Squared: 0.833,
Adjusted R-squared: 0.7495
F-statistic: 9.975 on 2 and 4 DF, p-value: 0.02789
> summary.aov(mod12)
Df Sum Sq Mean Sq F value Pr(>F)
x2
1 0.61820 0.61820 7.3403 0.05358 .
x1
1 1.06206 1.06206 12.6105 0.02377 *
Residuals
4 0.33688 0.08422
--Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` ' 1
The variation explained by x2 in model 11, the fit of x2 alone, (0.6182) is the same as in model
12, the fit of y on x2 and x1 (in that order).
Returning to model 12, we see that, in the presence of x2, x1 explains a further 1.0621 of the total
variation. Together the two variables explain 83.3% of the total variation (much more than the
30.6% explained by x2 alone). In the presence of x2, we gain a lot by including x1.
It is clearer and simpler if we fit the variable which explains most of the variation in the
responses first, then the variable of next importance and so on.