CHAPTER 5
advertisement
CHAPTER 5
The Evolution Trees
In biological research, it is often necessary to describe the relationship among species. If
we assume that these species all evolve from a common ancestor, then we like to
construct an evolution tree with this mysterious ancestor as the root and the species as the
leaf nodes. There will be internal nodes which represent unknown species and the length
of each edge (a, b) represents the time needed to evolve from a to b .
5.1 Rooted and Unrooted Evolution Trees
Let us first clarify the following points about evolution trees:
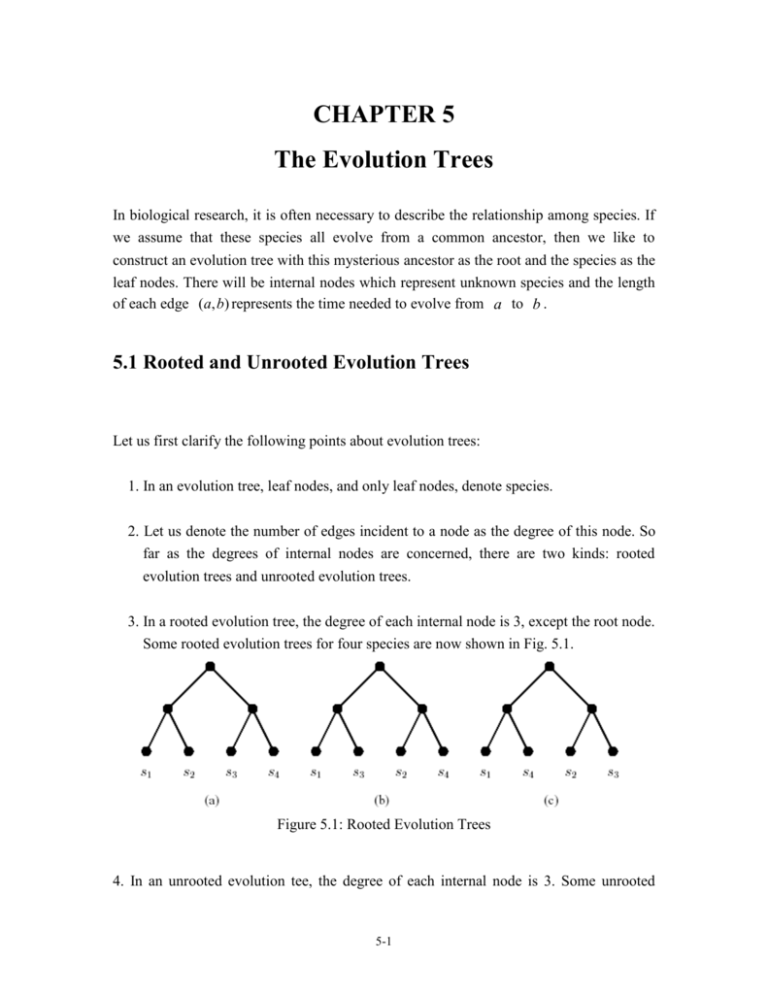
1. In an evolution tree, leaf nodes, and only leaf nodes, denote species.
2. Let us denote the number of edges incident to a node as the degree of this node. So
far as the degrees of internal nodes are concerned, there are two kinds: rooted
evolution trees and unrooted evolution trees.
3. In a rooted evolution tree, the degree of each internal node is 3, except the root node.
Some rooted evolution trees for four species are now shown in Fig. 5.1.
Figure 5.1: Rooted Evolution Trees
4. In an unrooted evolution tee, the degree of each internal node is 3. Some unrooted
5-1
evolution trees for four species are now shown in Fig. 5.2.
Figure 5.2: Unrooted Evolution Trees
5. We always assume that the input is a distance matrix among all of the species. Besides,
we always assume that the distances satisfy the triangular inequality relationship.
Depending upon different conditions, different evolution trees will be constructed to
reflect the distances among species.
6. If the evolution tree is rooted, then the distances from the root to all leaf nodes are the
same.
7. In every evolution tree, let dt ( si , s j ) denote the distance between species s i and s j . Let
d ( s i , s j ) denote the distance between s i and s j in the distance matrix. Then
dt ( si , s j ) d ( si , s j )
Let us now compare the number of rooted trees and the number of unrooted trees. First,
let us consider the unrooted trees.
5-2
Let NE (n) denote the number of edges of an unrooted evolution tree. It can be easily
seen that whenever a new species is added to an unrooted evolution tree, the number of
edges of the tree is increased by 2. It can also be proved by induction that the following is
true:
NE (n) 2n 3.
Given an unrooted evolution tree, we can add a new species into the evolution tree
by splitting any edge as shown in Fig 5.3. Let TU(n) denote the number of unrooted
evolution trees for n species. Since there are 2n 3 edges in an unrooted evolution tree
5-3
Figure 5.3: Inserting a New Species to an Unrooted Evolution Tree
with n species, we have
TU (n 1) (2n 3)TU (n),
or
TU (n) (2n 5)TU (n 1)
That is,
TU (n) (2n 5)( 2n 7) 1
We can now determine the number of rooted trees for n species. Given an unrooted
evolution tree for n species, we can convert it into a rooted evolution tree by splitting any
edge of the tree and adding a root node, as shown in Fig. 5.4.
Since there are 2n 3 edges in every unrooted evolution tree for n species, there
are
(2n 3)TU (n)
rooted evolution trees for n species. Let TR (n) denote the number of rooted trees for n
species. We have
TR (n) (2n 3)TU (n)
(2n 3)( 2n 5)( 2n 7) 1
TU (n 1).
5-4
Figure 5.4: Changing Unrooted Evolution Trees into Rooted Evolution Trees
This means that the number of rooted evolution trees is much higher than that of the
unrooted evolution trees. When n is very large, it will be desirable to consider unrooted
evolution trees. But, we can not explain evolution by an unrooted tree. What we can do is
to add a species which is exceedingly different from the species which we are analyzing.
This outlier species will cause a long link and we can use that to identify a root. Fig. 5.5(a)
5-5
shows such a case. Since s5 is so far away from the other species, we may discard it and
then say that we have obtained a rooted evolution tree as shown in Fig. 5.5(b).
Figure 5.5: An Unrooted Evolution Tree with an Outlier Species
5.2 Minimax, Minisum and Minisize Evolution Trees
In the previous section, we discussed the concept of rooted and unrooted evolution trees.
Note that the input of an evolution tree problem is a distance matrix and we are asked to
construct an evolution tree to properly reflect these distances. We need to specify
conditions under which an evolution tree can be built. The following different
specifications will give us different evolution trees. Note that dt ( si , s j ) (d ( si , s j ))
denotes the distance between s i and s j in the evolution tree (the distance between s i
and s j in the input distance matrix).
1. Minimax Evolution Trees
In a minimax evolution tree, the maximum of (dt ( si , s j ) d ( si , s j )) is minimized.
5-6
2. Minisum Evolution Trees
In a minisum evolution tree, the total sum of all pairs of distances among leaf nodes is
minimized. Thus this is very similar to a minimum routing cost tree except the distance
always refers to the distance between two leaf nodes.
3. Minisize Evolution Trees
In a minisize evolution tree, the total length of the tree is minimized.
In this chapter, we shall introduce a new approach to construct rooted volution trees. It
is called the minimal spanning tree approach. It has been found that the rooted and
unrooted minisum evolution tree problems are all NP-complete. It has also been found
that the unrooted minimax evolution tree problem and the rooted minisize evolution tree
problem are NP-complete. Whether the unrooted minisize evolution tree problem is
NP-complete or not is still an open problem. Finally, the rooted minimax evolution tree
problem has a polynomial algorithm. We will introduce this algorithm in the next
section. Table 5.1 summarizes the above statements.
Table 5.1: The Complexities of Evolution Tree Problems
5.3 A Minimax Rooted Evolution Tree Algorithm
In this section, we shall introduce a minimax evolution tree algorithm for rooted
evolution trees. The algorithm which we are going to introduce is based upon the minimal
spanning tree concept. Consider Fig. 5.6. There is a minimal spanning tree in this figure.
Note that the edge (b, e) is the longest. If we break this edge, we obtain two subtrees. In
each subtree, suppose that x and y are nodes in the same subtree. Then the distance
between x and y is always smaller than the length of (b, e) . The property of minimal
spanning trees can be used to produce an unrooted minimax evolution tree.
5-7
Figure 5.6: A Minimal Spanning Tree
Our algorithm is a recursive one. Its basic principle is as follows. Let s i and s j be the
two species which have the longest distance in the distance matrix. Then our rooted
minimax evolution tree will have two subtrees as shown in Fig. 5.7. Subtree Ti contains
s i and T j contains s j . Besides, the distance from the root to leaf node s i is equal to the
1
distance from the root to leaf node s j . Both distances are equal to d ( s i , s j ) . That is, we
2
make sure that this longest distance is exactly preserved.
Figure 5.7: The Rooted Minimax Evolution Tree Based upon s i and s j where d ( s i , s j ) is
the largest and dt ( si , s j ) d ( si , s j )
There are two problems:
5-8
1. How are subtrees Ti and T j obtained? They are obtained recursively. That is, we apply
this algorithm to find these two subtrees.
2. What is the mechanism to determine which species are in Ti and which species are in
T j ? We will elaborate this in the following paragraphs.
We construct a minimal spanning tree based upon the distance matrix. Consider the
path linking s i and s j on this minimal spanning tree. Let e be the longest edge of this path.
If we delete this edge, we will obtain two sets of nodes. One subset of nodes will be in
Ti and the other subset of nodes will be T j .
The following is the algorithm.
Algorithm 5.1 A Rooted Minimax Evolution Tree Algorithm.
Input: A Distance Matrix of a Set S of nSpecies s1 , s2 ,, s n .
Output: A Rooted Minimax Evolution Tree for S .
Step 1: If S contains only one species x , return node x as the tree.
Step 2: Find the longest d ( si , s j ) in the distance matrix. Find a minimal spanning tree of
S.
Step 3: Find the longest edge e in the path linking si and s j in the minimal spanning tree.
Let S i and S j be the two sets of species obtained by breaking edge e .
Step 4: Use this algorithm recursively to find subtrees Ti and T j for S i and S j respectively.
Step 5: Construct a rooted tree with Ti and T j as subtrees. Let the distance from the root
r of this tree to the root of Ti (T j ) be hi (h j ). Set hi ( h j ) so that
dt (r , si ) dt (r , s j )
5-9
1
d ( si , s j )
2
Let us consider an example of the distance matrix in Table 5.2.
Table 5.2: A Distance Matrix
Our algorithm proceeds as follows:
1. The distance between s2 and s4 is the longest.
2. We construct a minimal spanning based upon the distance matrix as shown in Fig. 5.8.
Figure 5.8: A Minimal Spanning Tree Based upon Table 5.2
3. In the minimal spanning tree, the path linking s2 and s4 is ( s2 , s1 , s3 , s4 ). The longest
edge is ( s1 , s3 ) . Breaking this edge, we obtain two subsets of species :
S2 (s1 , s2 ) and S 4 (s3 , s4 ).
4. We construct two subtrees T2 and T4 for S 2 and S 4 respectively as shown in Fig. 5.9. Note
that in T2 (T4 ) , the distance dt (s1 , s2 ) d (s1 , s2 ) (dt ( s3 , s 4 ) d ( s3 , s 4 ))
5-10
Figure 5.9: Subtrees for T2 and T4
5. We combine these two subtrees by making sure that dt (s2 , s4 ) d (s2 , s4 ) as
shown in Fig. 5.10.
Figure 5.10: A Rooted Minimax Evolution Tree Based upon Table 5.2
Let us give another example so that the reader can have more feeling about the
algorithm. Suppose that we only consider s1 , s2 and s 3 in Table 5.2. Then the
corresponding rooted minimax evolution tree will look like that in Fig. 5.11. In this case,
the distance between s1 and s 3 and the distance between s1 and s 3 are both exactly preserved.
The distance between s1 and s 3 is preserved because this is the longest one to begin with. If
we break the longest edge in the path linking s1 and s 3 , we obtain a subset consisting
of s1 and s2 . This is why the distance between s1 and s2 is also preserved.
5-11
Figure 5.11: A Minimax Rooted Evolution Tree Based upon s1, s2 and s3 in Table 5.2.
5.4 The Determination of Weights When the Evolution Tree
Structure Is Given
In the above section, we showed that there are different criteria for constructing evolution
trees. Let us consider the case where we are asked to construct an unrooted evolution tree
for four species, namely s1 , s 2 , s3 and s4 . Let d ij denote the distance between s i and s j .
One possible such evolution tree is shown in Fig. 5.12.
Figure 5.12: A Possible Unrooted Evolution Tree for Four Species.
Let us further assume that we require our unrooted evolution tree to be a minisize
evolution tree and the tree shown in Fig. 5.12 is the best one for this purpose. Then we
can determine xi ’s for i 1 to 5, by the linear programming approach as follows:
5-12
Minimize
x1 x2 x3 x4 x5
Suppose that our evolution tree is a rooted one, and somehow, we have determined
that the tree in Fig. 5.13 is the desired one. Then we will have the following equations for
the linear programming problem:
5-13
Figure 5.13: A Possible Rooted Evolution Tree for Four Species
If some other criteria is used for constructing our tree, then different linear
programming equations will be used. This approach cannot be used for minimax
evolution tree for unrooted evolution trees because it is unknown how to formulate this
problem as a linear programming problem.
The problem is how to determine the structure of the tree. As explained in Section 5.1,
the number of possible evolution trees is exponential with respect to n . We can not
exhaustively find every possible evolution tree and apply the linear programming
technique. In the following sections, we will introduce two heuristic algorithms to
5-14
determine a reasonably good structure of evolution trees for a given input distance
matrix.
5.5 The Unweighted Pair Group Method with Arithmetic
Mean (UPGMA) for Rooted Evolution Trees
The unweighted pair group method with arithmetic mean (UPGMA) is a method to
produce a good rooted evolution tree after a distance matrix is given. Let us consider the
distance matrix in Table 5.3:
Table 5.3: A Distance Matrix
Our method is in the spirit of the greedy method.
Step 1. Select the pair of species with the smallest distance between them. s 3 and s4 are
selected. Construct a rooted evolution tree with s 3 and s4 as leaf nodes, as shown in
Fig. 5.14.
Step 2. Consider ( s3 , s 4 ) as a new specie. The new distances are updated as follows:
5-15
Figure 5.14: A Rooted Evolution Tree with s 3 and s4 as Leaf Nodes
The new distance matrix will be shown in Table 5.4.
Table 5.4: A New Distance Matrix
Since d ( s1 , ( s3 , s 4 )) is the smallest, we select s1 and ( s3 , s 4 ) Construct a rooted
evolution tree as that shown in Fig. 5.15.
Figure 5.15: The Rooted Evolution Tree with s1 Added
5-16
Step 3. Since s4 is the only specie left, the final tree will look like that shown in Fig. 5.16.
Figure 5.16: The Final Evolution Tree Constructed by UPGMA
This evolution tree structure is a heuristic solution for rooted evolution tree problems.
After obtaining this structure, we will can then use the linear programming technique to
produce an evolution tree for a given criteria.
In the following, the algorithm for UPGMA is given.
Algorithm 5.2 The Unweighted Pair Group Method with Arithmetic Mean Algorithm.
Input: A set S of n species and its distance matrix.
Output: A rooted evolutionary tree structure for S .
Step 1: Find two species x and y such that d ( x, y ) is the shortest.
Step 2: Create a new species, denoted as ( x, y ) .
Construct a tree using ( x, y ) as the root and subtrees rooted at x and y respectively
as the descendants of the root ( x, y ) .
Delete x and y from the distance matrix.
Step 3: If all species have been deleted,
return the tree rooted at ( x, y ) and exit.
Otherwise update the distance to a new distance matrix.
The distance d ( z, ( x, y )) is calculated as:
5-17
d ( z, ( x, y ))
1
(d ( z , x)) d ( z, y ))
2
Step 4: Go to Step 1.
5.6 The Neighbor Joining Method for Unrooted Evolution
Trees
The neighbor joining method is a method to produce a reasonably good unrooted
evolution tree structure. Let us first give an example. Consider the same distance matrix
as given in Table 5.3 in the above section. The neighbor joining method proceeds as
follows:
We first construct a 1-star, shown in Fig. 5.17 with species as leaf nodes. The distance
from the unique internal node to a leaf node is the mean of the distances from this specie
to all other species. For instance, the weight of the edge ( x, s1 ) is:
1
(d ( s1 , s2 ) d ( s1 , s3 ) d ( s1 , s4 ))
3
1
(4 4 3)
3
3.67
W ( x, s1 )
In the following, let average( s i ) =
1
i j d (si , s j ). In the present connection,
n 1
W ( si , x) average( s i ).
The tree in Fig. 5.17 is not an unrooted evolution tree. To determine the structure of an
unrooted evolution tree, we have to determine which two species are to be paired.
5-18
Figure 5.17: A 1-Star to Initiate the Neighbor Joining Method
Let us now imagine that s1 and s2 are to be paired. At present, s1 and s2 are connected as
shown in Fig.5.18(a). Our job is to insert an internal node to that in Fig.5.18(a) as shown
in Fig. 5.18(b).
Figure 5.18: The Connection of s1 and s2
If we consider the three nodes, namely s1 , s2 and x as a triangle, where the
edge (s1 , s2 ) has weight equal to d (s1 , s2 ) = 4, we may set node x1 as the geometrical
center of this triangle. The new connection cost NC is as follows:
The weights of edges (s1 , x1 ) , (s2 , x1 ) and ( x1 , x) in the new connection are calculated as
follows:
5-19
Note that W (s1 , x1 ) W (s2 , x1 ) W ( x1 , x) is equal to the new connection cost. This
can be proved as follows:
Therefore,
This means that if we pair s1 and s2, we would have a structure as shown in Fig. 5.19. The
old connection cost OC is (average( s1 )+average( s2 )) = 5 + 3.67 = 8.67. The new
connection cost is NC = 1.33 + 2.67 + 2.33 = 6.33. The cost saved is OC NC = 8.67 6.33 = 2.34. Besides,
5-20
Figure 5.19: The Structure Pairing s1 with
Thus in the new connections, the distance between s1 and s2 is exactly preserved.
In fact, the cost saved is equal to (average( s1 ) + average( s2 ))
average( s2 ) + d (s1 , s2 ) )
1
(average( s1 ) +
2
1
( average( s1 )+average( s2 ) d (s1 , s2 ) ).
2
Through the same mechanism, we can try to pair s1 with s3. The old and new structures
are shown in Fig. 5.20.
Figure 5.20: The Pairing of s1 with s3
The new connection cost is
New connection cost = (3.67 4 4) / 2 5.835. (3.67 + 4 + 4)/2 = 5.835. The
old connection cost is 3.67 4 7.67. The cost saved is 7.67 5.835 1.835 Using the
same technique, we can find the following:
The cost saved by pairing s1 with s4 is 2. The cost saved by pairing s2 with s 3 is 1.5. The
cost saved by pairing s2 with s4 is 1.67. The cost saved by pairing s 3 with s4 is 2.67.
We conclude that the pairing of s 3 and s4 produces the largest cost saving. Thus we
pair s 3 with s4 and we have the structure as shown in Fig. 5.21.
5-21
We can now apply the linear programming technique to this structure. The linear
programming equations will depend upon which criteria we use.
The algorithm of neighbor joining method is now presented as follows:
Figure 5.21: The Final Tree Structure Resulting from Pairing s 3 and s4
Algorithm 5.3 Neighbor joining method.
Input: A set S of n species and its distance matrix.
Output: An unrooted evolutionary tree structure for S.
Step 1: Construct a 1-star tree T with x as center node and species as leaf nodes.
Calculate average( s i ) =
1
n 1
j i
d ( si , s j ).
k = 1.
Step 2: If the degree of x is greater than 3, .nd two species si and sj adjacent to x such that
(average( s i ) + average( s j ) - d ( s i , s j ) ) is maximized.
Step 3: Insert an interval node x k with degree 3 into T such that x k is connected to x, s i
and s j .
Step 4: If the degree of x is equal to 3, return T and exit; otherwise k = k + 1 and go to
Step 2.
5-22
5.7 An Approximation Algorithm for an Unrooted
Minisize Evolution Tree
In Section 5.2, we introduced the concept of minisize unrooted evolution trees and we
indicated that we have not found any polynomial algorithm for this problem, neither have
we proved that this problem is NP-complete. In this section, we will introduce an
approximation algorithm for this problem. We will also show that the size of this
approximate solution is never larger than twice of the size of an optimal solution. Thus
the error rate is 1 for this approximation algorithm.
This algorithm is based upon the minimal spanning tree. Basically, it transforms a
minimal spanning tree constructed out of a distance matrix into an unrooted evolution
tree while maintaining its total length.
Let us consider Table 5.3 again. We first construct a minimal spanning tree out of this
distance matrix. This minimal spanning tree is now shown in Fig. 5.22.
Figure 5.22: A Minimal Spanning Tree Based upon the Distance Matrix in Table 5.3.
After obtaining this minimal spanning tree, we can transform it into an unrooted
evolution tree as shown in Fig. 5.23.
5-23
Figure 5.22: A Minimal Spanning Tree Based upon the Distance Matrix in Table 5.3.
Let us now explain how the unrooted evolution tree is obtained. Given a tree, we can
order the nodes in the tree through a breadth first search. Consider the
Figure 5.23: An Unrooted Evolution Tree Transformed from the Minimal Spanning Tree
in Fig. 5.22
The breadth first search would start from the root and visit all of the first level
descendants first, then the second level descendants, and so on. For the tree shown in Fig.
5.24(b), a breadth first search would give the following order of the nodes: e, b, g, j, f, a,
c, d, h, i. For the tree in Fig. 5.22, if we use s4 as the root, we would have the following
order of nodes: s 4 , s3 , s1 , s 2 .
Once we conduct a breadth first search of the minimal spanning tree, we simply
5-24
transform the minimal spanning tree into an evolution tree by adding nodes one by one,
through the ordering obtained by the breadth first search.
1. We first start by linking s 3 to s4 . The weight of the edge linking s4 and s3 will be the
same as that in the minimal spanning tree. This results in a simple unrooted evolution
tree as shown in Fig. 5.25(a).
2. We then link s1 with s4 . We can not link these two directly because this will make
s4 an internal node with degree 2, which is not allowed in an unrooted evolution tree.
We therefore create a new node x1 on the edge emanating from s4 . We link s1 to x1 .
The weight of ( x, s1 ) is set to be equal to that of (s4 , s1 ) in the minimal spanning tree.
The weights of ( x1 , s4 ) and ( s3 , x1 ) are set to be 0 and 2 respectively in the purpose of
keeping the total length of the minimal spanning tree. This is shown in Fig. 5.25(b)
Figure 5.25: The Adding of Nodes to Form an Unrooted Evolution Tree
3. The other species are added to the partially constructed unrooted evolution tree one by
one with the same procedure.
Let us summarize our unrooted evolution tree construction as follows:
Algorithm 5.4 An Approximation Algorithm for an Unrooted Minisize Evolution Tree
whose Error Rate Is 1.
Step 1: Construct a minimal spanning tree based upon the given distance matrix.
Step 2: Conduct a breadth first search on this minimal spanning tree. Without losing
generality, we may say that the nodes are ordered as s1 , s2 ,, s n .
Step 3: Add species one by one to form an unrooted evolution tree. The rules of adding
species are as follows:
5-25
(a) If there is only one species in the partially constructed evolution tree, link the
new specie directly to it.
(b) If the partially constructed evolution tree contains more than one specie and
our procedure requires us to link si 1 to s i . Create a new internal node x in the
edge emanating from s i . Link si 1 to x. Let the weight of ( x, si ) be 0 and the
weight of ( si , si 1 ) be the weight of ( si , si 1 ) in the minimal spanning tree. Let
the weight of ( x, si 1 ) be the weight of ( si , si 1 ) in the minimal spanning tree.
We can see that the evolution tree in Fig. 5.23 is obtained by applying the above rules
to the minimal spanning tree. Of course, we have to prove that this tree is indeed an
evolution tree. The degree of each internal node is exactly three. The only thing that we
have to prove is that dt ( si , s j ) d ( si , s j ). This is true because the distance between any
two species on the evolution tree is exactly the same as that on the minimal spanning tree.
Yet the distance between any two species on the minimal spanning tree must be larger or
equal to the distance between them in the distance matrix because of the triangular
inequality. Therefore, we can be assured that the distance between any two species on the
constructed unrooted evolution tree is larger or equal to the distance between them in the
distance matrix.
In the following, we will prove that the total length of this unrooted evolution tree is
less than or equal to twice of the length of an optimal unrooted minisize evolution tree.
We first have to introduce a very important concept: the Hamiltonian cycle. Given a
graph G = (V,E), a Hamiltonian cycle is a cycle visiting all of the nodes exactly once. For
instance, consider the graph in Fig. 5.26. The cycle a b d c e a is a Hamiltonian
cycle. In a graph, there may be several Hamiltonian cycles and there may be no
Hamiltonian cycle at all. The traveling salesperson problem is to find a Hamiltonian cycle
with the smallest length. This problem has been found to be NP-complete.
Figure 5.26: A Graph to Illustrate Hamiltonian Cycles
5-26
Given a graph, let TSP denote an optimal solution of the traveling salesperson problem
and MST denote a minimal spanning tree of the graph. Let us now prove that the length of
MST is smaller than that of TSP. This can be done as follows: Delete any edge from the
TSP. Let the resulting graph be denoted as P. P is a spanning tree. Obviously the length
of P is smaller than that of TSP. Furthermore, the length of MST is smaller than or equal
to that of P because MST is the smallest spanning tree. Thus we can conclude that the
length of MST is smaller than that of TSP. That is,
We are given a distance matrix to start with. Based upon this distance matrix, we can
construct a complete graph out of this distance matrix. A complete graph is a graph where
every pair of nodes are connected. For instance, the complete graph corresponding to the
distance matrix in Table 5.3 is now shown in Fig. 5.27(a). A TSP of this graph is shown
in Fig. 5.27(b). We can see that the length of the minimal spanning tree in Fig. 5.22,
which is 9, is smaller than that of the TSP, which is 15.
Figure 5.27: The Complete Graph of the Distance Matrix in Table 5.3 and a TSP
Note that our constructed unrooted evolution tree has the same length as that of the
minimal spanning tree of the complete graph. We conclude that the length of our
constructed unrooted evolution tree is smaller than that of the TSP of the complete graph
constructed out of the input distance matrix. Let the size of the tree thus constructed be
denoted as APP. Then
5-27
In the following, we will prove that the length of the TSP is never larger than twice of
the length of an optimal unrooted minisize evolution tree. To do this, we will have to
introduce a term, called Euler tour. Given a graph, an Euler tour is a cycle which
traverses each edge exactly once. For instance, for the graph shown in Fig. 5.28, the cycle
a b c d b e a is an Euler tour. Note that an Euler tour may visit a node more
than once. Again, not every graph has an Euler tour. For instance, there is no Euler tour in
the graph of Fig. 5.26.
Figure 5.28: A Graph to Illustrate Euler Tours
It can be easily seen that there is no Euler tour in any tree. But, if we duplicate every
edge of a tree, there is an Euler tour in this resulting graph. For instance, consider the
evolution tree in Fig. 5.23. The result of duplicating every edge of it is shown in Fig. 5.29.
It is obvious that there is an Euler tour in this graph. For instance, one of them is the
following cycle:
which corresponds the following cycle of species
5-28
Figure 5.29: The Result of Duplicating Every Edge in the Tree of Fig. 5.23.
Let OPT denote an optimal unrooted minisize evolution tree T. Let ET denote any
Euler tour of the graph obtained by duplicating every edge of this optimal tree T. Then
|ET| = 2|OPT| . Without losing generality, we may say that :
|ET| = dt ( s1 , s 2 ) dt ( s 2 , s3 ) dt ( s n1 , s n )
where dt ( si , s j ) denotes the distance between s i and s j in T.
We shall prove that |TSP| is smaller than or equal to |ET|. This can be seen as follows.
Consider the cycle of species corresponding to the Euler tour of the duplicated tree. Let
this cycle be denoted as CET. In our case, for instance, CET is s1 s4 s3 s 2 s1 . In a
general case, without losing generality, we may say that this cycle is s1 s 2 s n .
The total length of this cycle is d ( s1 ,s 2 ) d ( s 2 , s3 ) d ( s n1 , s n ) where d ( s i , s j )
denotes the distance between s i and s j in the distance matrix. Since T is an evolution tree,
dt ( si , s j ) d ( s i , s j ) for all s i and s j . Therefore, |CET| |ET|.
Note that TSP is the shortest Hamiltonian cycle of the complete graph constructed out
of the distance matrix. CET is also a Hamiltonian cycle of the complete graph. Therefore,
|TSP| |CET| |ET| = 2|OPT|.
But, we proved previously that APP = |MST| < |TSP|. Consequently, we have
5-29
5.8 The Minimal Spanning Tree Preservation Approach
for Evolution Tree Construction
Let D and Dt denote the original input distance matrix and the distance matrix based upon
the evolution tree. Let MST(D) (MST(Dt)) denote the minimal spanning tree constructed
out of distance matrix D (Dt). The condition for our minimal spanning tree approach for
the evolution tree construction problem is that MST(D) is an MST(Dt).
Basically, given an input distance matrix D, we first construct a minimal spanning tree
MST(D) out of it. We then sort the edges of MST(D) into an ascending sequence. We then
consider edges from the smallest one by one. For each edge, we add a new internal node
into the partially constructed evolution tree. If there is one node connected by this edge
which is not yet in the partially constructed evolution tree, we also add this node to the
tree as a leave node.
We present the minimal spanning tree preservation approach for the evolution tree
construction in Algorithm 5.5.
Algorithm 5.5 A Minimal Spanning Tree Preservation Approach for the Evolution Tree
Construction.
Input: A distance matrix D(n, n) for a set S of n species.
Output: A rooted evolution tree for S such that MST(D) is equal to one of MST( Dt ).
Step 1: Find a minimal spanning tree MST(D) of D.
Step 2: Sort the edges of the spanning tree by their weights in the ascending order.
Let the result be e1 , e2 ,,e n1 , where | e i | < | e j | if i < j.
Step 3: Create a leaf node for each species.
Step 4: for k = 1 to n 1 do
Let the two species connected by ek be s k1 and s k 2 .
Construct a new internal N k with descendants Tk1 (the subtree containing
s k1 ) and Tk 2 (the subtree containing s k 2 ) such that :
5-30
dt ( N k , sk1 ) dt ( N k , sk2 )
1
max{ d ( s ' k1 , s ' k2 ) | s ' k1 and s ' k2 are species inT k 2 and Tk2 , respective ly}.
2
end for
Step 5: Output the evolution tree.
We illustrate the idea of Algorithm 5.5 by the following example.
Consider the distance matrix D in Table 5.5. An MST(D) is illustrated in Figure 5.30.
Table 5.5: A New Distance Matrix
Figure 5.30: A Minimal Spanning Tree Constructed out of Table 5.5
The edge sequence sorted by the edges’weights on MST(D) in the ascending order is
e(4, 5), e(1, 2), e(2, 3), e(5, 6), e(3, 4) ( d(4, 5) = 2, d(1, 2) = 3, d(2, 3) = 4, d(5, 6) = 5,
d(3, 4) = 7 ). Then n leaves are constructed to represent the input n species. Let us
consider the smallest edge e(4, 5) on MST(D). We add a new internal node N1 with
5-31
1
d (4,5) 1 . This ensures
2
dt (4,5) d (4,5) as desired. Obviously, MST(D) of species 4 and 5 is MST( Dt ) of species
descendants 4 and 5 as below. Note that dt ( N 1 ,4) dt ( N 1 ,5)
4 and 5.
For the second smallest edge e(1,2) , a new internal node N 2 with descendants 1 and 2
are constructed as below with dt ( N 2 ,1) dt ( N 2 ,2)
1
d (1,2) 1.5 .
2
For the third smallest edge e( 2,3) , a new internal node N 3 with descendants 3 and the
subtree T1 which contains species 2 is constructed as below with
dt ( N 3 ,2) dt ( N 3 ,3) max{ d (1,3), d (,2,3)} 3.5 . Again, the MST(D) of species 1, 2 and
3 will be an MST( Dt ) of species 1, 2 and 3.
Likewise, for the fourth smallest edge e(5,6) , we construct a new internal node N 4 as
below with dt ( N 4 ,5) dt ( N 4 ,6) max{ d (4,6), d (,5,6)} 2.7
5-32
For the last edge e(3, 4), a new internal node N5 is constructed with dt(N5, 3) = dt( N 5 , 4)
= max{ d (s ' k1 , s ' k2 ) | s ' k1 {1,2,3} and s ' k2 {4,5,6}} d (1,6) 8.4. The final evolution
tree is shown in Figure 5.31. The dt-matrix corresponding to this evolution tree is shown
in Table 5.6. Based upon this dt-matrix, we can construct several distinct minimal
spanning trees and one of them is shown in Figure 5.32 which is exactly the same as that
in Figure 5.30 except the weights of the edges are not the same any more. Thus, this
evolution tree satisfies the minimal spanning tree preservation approach criterion.
Figure 5.31: An Evolution Tree Based on the Distance Matrix in Table 5.5
Table 5.6: The Distance Matrix Dt Based on the Evolution Tree in Fig 5.31
5-33
We shall prove the minimal spanning tree preservation property of our Algorithm 5.5.Our
proof is based upon a special property of minimal spanning trees: Given a spanning tree T
for a set of nodes S, T is a minimal spanning tree if every edge e(a, b) satisfies
Figure 5.32: A Minimal Spanning Tree Based on the Distance Matrix in Table 5.6
Condition 5.1 as follows.
Condition 5.1 Suppose we break the edge e(a, b) on T. The set S is split into two subsets
S a and S b containing a and b respectively. Then the distance between any node a in
S a and any node b_ in S b is not smaller than the distance between a and b.
Before the formal proof, let us introduce the concept of the lowest common ancestor in
an evolution tree. In a rooted evolution tree T, the lowest common ancestor of two species
x and y, denoted as lca ( x, y ) , is the deepest internal node in T that is an ancestorof both x
and y. From Algorithm 5.5, we know that internal node Ni corresponds to an edge
e( si1 , si2 ) on MST(D) and N i is exactly the lowest common ancestor of s i1 and s i2
on the evolution tree. Thus, we have lca ( si1 , si2 ) . Since Algorithm 5.5 constructs an
evolution tree from bottom to top, we define the level of a node x, denoted as level(x), in
an evolution tree as follows: For each leave node x, level(x) = 0. Let a node x be an
immediate ancestor of node y, then level(x) = level(y) + l. Let a ' and b ' be connected by
a path including e(a, b) in MST(D). Based upon these definitions, we observe the
following facts:
Fact 5.1 If nodes a, a, b and b are included in a partially constructed evolution tree
produced by Algorithm 5.5 and level(lca(a, b)) level(lca( a ' , b ' )), dt(a, b) dt( a ' , b ' ).
5-34
Fact 5.2 During the execution of Algorithm 5.5, suppose node a is in a partially
constructed evolution tree Ta and node b is not in Ta. Let Ra denote the root of Ta. When
node b is connected to Ta so that a and b are in the same connected component Tab. Let
Rab denote the root of Tab. Then level(Ra) level(Rab).
Fact 5.3 During the execution of Algorithm 5.5, every partially constructed evolution tree
corresponds to a connected component of MST(D).
Lemma 5.1 Let a_ and b_ be connected by a path including e(a, b) in MST(D). Then
dt( a ' , b ' ) dt(a, b) by Algorithm 5.5.
Proof. When e(a, b) is considered, there are three cases:
Case 1. Neither a nor b is in any partially constructed evolution tree. In this case, Ni =
lca(a, b) is the immediate ancestor of a and b as shown in Figure 5.33. Thus,
level(lca( a ' , b ' )) must be larger than level(lca(a, b)).
Figure 5.33: A Partially Constructed Evolution Corresponding to Case 1
Case 2. Only one of a and b is in a partially constructed evolution tree. Without losing
generality, we may assume that a is in some partially constructed evolution tree as shown
in Figure 5.34 (a). Now, e(a, b) is considered. Thus, b will be connected to the subtree
containing a, as shown in Figure 5.34 (b). Later, when some edge is considered such that
this consideration puts a, b and b_ into a connected component, denoted as T, as shown in
Figure 5.34 (c). If a ' is already in this evolution tree T, a ' must be in Ta according to
Fact 5.3 and lca( a ' , b ' ) will be the root of T, denoted as RT . Thus level(lca(a, b)) <
level(lca( a ' , b ' )). If a ' has not appeared yet, according to Fact 5.2,
5-35
Figure 5.34: A Partially Constructed Evolution Tree Corresponding to Case 2
level(lca( a ' , b ' )) will be even higher than level( RT ). This means that level(lca(a, b)) will
be smaller than level(lca( a ' , b ' )).
Case 3. Nodes a and b exist in two different partially constructed evolution trees, as
shown in Figure 5.35 (a). Since e(a, b) is now considered, these two partially constructed
evolution trees will be connected into a connected component as shown in Figure 5.35 (b).
Thus lca(a, b) is the root of the connected component and its level is the highest. If both
a ' and b ' are in this tree, a ' will be in the subtree containing a and b ' will be in the subtree
containing b according to Fact 5.3. Thus lca(a, b) = lca( a ' , b ' ). If at least one of a ' or
b ' is not in this connected component, the level of lca(a, b) will be smaller than the level
of lca( a ' , b ' ), according to Fact 5.2.
Let us replace the (n 1) weights on MST(D) by the corresponding (n 1) distances
on Dt constructed by Algorithm 5.5. Let the new structure be Tt . We shall show that Tt is
an MST(Dt).
Theorem 5.1 Tt is an MST( Dt ).
Proof. Suppose some edge, say e(a, b), on Tt is broken. Let the two split subtrees be Ta
(containing a) and Tb (containing b). The path from any species a ' in S a to any species
5-36
Figure 5.35: A Partially Constructed Evolution Tree Corresponding to Case
b ' in S b must include e(a, b) on MST(D). From Lemma 5.1, dt( a ' , b ' ) dt(a, b). That is,
every edge on Tt which is a spanning tree of Dt satisfies Condition 5.1. Therefore Tt is an
MST( Dt ).
In fact, the minima tree algorithm, Algorithm 5.1, introduced in the above section also
meets the minimal spanning tree preservation criteria.
From Algorithm 5.1, we know that each internal node in the evolution tree is based
upon the longest edge e in the path linking s i and s j in MST(D) where D( s i , s j ) is the
longest distance among species currently considered, namely S. Let S be divided into two
subsets S i (containing s i ) and S j (containing s j ) after e is broken in MST(D). Then
dt (s ' i , s ' j ) d (si , s j ) where si s ( s j s ) are species in S i ( S j ). Algorithm 5.1 recursively
'
'
finds subtrees Ti and T j for S i and S j respectively. Note that since d ( s i , s j ) is the longest
distance among species in S ( S i S j ) ,
dt (s ' i , s ' j ) d (si , s j ) dt (s ' i1 , s ' i 2 )
and
dt ( s i , s ' j ) dt ( s ' j1 , s ' j 2 ) where s ' i1 ' s and s ' i 2 ' s ( s ' j1 ' s and s ' j 2 ' s ) are species in
'
S i (S j ) .
Based upon the statements that we prove the minimal spanning tree preservation for
5-37
Algorithm 5.5, it is easy to see that Algorithm 5.1 also preserves the minimal spanning
tree.
5-38