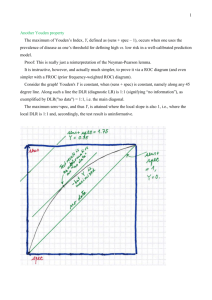
Incidence: The number of new cases of illness commencing, or of
advertisement

WP3 Food quality and safety T1 On farm risk analysis Dairy Farm Risk Analysis 2 Basic epidemiological measures used in summarising animals health monitoring data [E-J 1] The disease incidence is defined as number of new cases of illness commencing, or of persons falling ill, during a specified time period in a given population. In animal husbandry, where the animals disease status is usually monitored during long time period and one observation corresponds to the animal’s disease status in unit period, the disease incidence can be defined also as the number of observations with disease. For example the results about disease incidences in Estonian dairy cows are presented in Table 1. Note, that here the total number of disease incidents does not mean the number of different diseased cows, but this number shows the number of months where the cows had a certain disease summed over whole time period and studied farms. Table 1. Disease incidence Disease or disease group Disease incidence Udder diseases 4708 Uterine infection 2389 Metabolic diseases 1190 Retained placenta 751 Foot diseases 349 Other injuries 207 Enteritis 164 Disorders of rumen or abomasum 140 Ovulatory dysfunction 137 Dystocia 129 Abortion 50 Prolapse of uterus 52 Skin diseases 29 Diseases of respiratory tract 17 TOTAL 10312 Based on disease incidences the proportions of certain diseases in whole number of disease incidents can be found. For example the panorama of multifactorial diseases in Estonian dairy cows is given in Figure 1. The most common diseases of dairy cows in Estonia are udder diseases (45.7%), uterine infection (23.2%), metabolic diseases (11.5%), retained placenta (7.3%) followed by foot diseases (3.0%), other injuries (2.0%), enteritis (1.6%), disorders of rumen or abomasum (1.4%), ovulatory dysfunction (1.3%) and dystocia (1.3%). Relatively fewer cases were registered on abortion (0.5%), prolapse of the uterus (0.5%), skin diseases (0.3%) and diseases of the respiratory tract (0.2%). Figure 1. The panorama of multifactorial diseases in Estonian dairy cows Uterine infection 23.2% Metabolic diseases 11.5% Retained placenta 7.3% Foot diseases 3.4% Other injuries 2.0% Enteritis 1.6% Disorders of rumen or abomasum 1.4% Ovulatory dysfunction 1.3% Abortion 0.5% Udder diseases 45.7% Diseases of respiratory tract 0.2% Dystocia 1.3% Prolapse of uterus 0.5% Skin diseases 0.3% The prevalence rate (PR) is defined as the proportion of diseased animals in fixed time moment or period to population size in that moment or period. The incidence rate (IR) is defined as the proportion of new cases of illness commencing during a specified time period in a given population to the population at risk (the total number of observations). The estimate of where the true value of a result lies is usually expressed in terms of a 95% confidence interval (CI), or confidence limits. The calculation of confidence limits bases on the (asymptotic) distribution of studied characteristic. In case of large sample, usually the normal distribution is used to get approximate confidence intervals. For incidence rate the asymptotic confidence limits can be found using the following formula: 95% CIIR IR 1.96 IR (number of observations) . For example, the disease incidences, incidence rates in percents (multiplied with 100%) and the confidence intervals of incidence rates in Estonian milking cow’s study are presented in Table 2. The total number of observations was 87332. Table 2. Disease incidences in Estonian dairy cows (n = 87332) Disease or disease group Disease incidence Incidence rate (%) Udder diseases 4708 5.39 Uterine infection 2389 2.74 Metabolic diseases 1190 1.36 Retained placenta 751 0.86 Foot diseases 349 0.40 Other injuries 207 0.24 Enteritis 164 0.19 Disorders of rumen or abomasum 140 0.16 Ovulatory dysfunction 137 0.15 Dystocia 129 0.15 Abortion 50 0.06 Prolapse of uterus 52 0.06 Skin diseases 29 0.03 Diseases of respiratory tract 17 0.02 TOTAL 10312 11.81 95% CI 5.37–5.41 2.73–2.75 1.35–1.37 0.85–0.87 0.40–0.40 0.24–0.24 0.19–0.19 0.16–0.16 0.15–0.15 0.15–0.15 0.06–0.06 0.06–0.06 0.03–0.03 0.02–0.02 11.78–11.82 To compare the disease status in different groups the measure called as relative risk (RR) is used. The relative risk is defined as the ratio of the probability of developing, in a specified period of time, an outcome among those receiving the treatment of interest or exposed to a risk factor, compared with the probability of developing the outcome if the risk factor or intervention is not present. The RR can be calculated as the ratio of incidence rates. For example, comparing the keeping conditions in Estonian milking cow’s study, there were in total 6600 observations with free stall keeping and 80732 observations with tie keeping. There were registered 190 and 4518 udder diseases, respectively in case of free stall keeping and tie keeping. The corresponding incidence rates are 190/ 6600 0.0288 and 4518/ 80732 0.0560 . The risk for cow to get an udder disease in case of tie keeping compared to free stall keeping is estimated as 0.0560/ 0.0288 1.94 . For relative risk the asymptotic confidence limits can be found using the following formula: 95% CIRR eln(RR)1.96se[ln(RR)] e RR 1.96se[ln(RR)] ; RR e1.96se[ln(RR)] , where ln is the natural logarithm, e is the known constant (e = 2.71828… ); se[ln(RR)] 1 1 number of cases in exposed animals number of cases in non-exposed animals and se denotes the standard error. For example, comparing the keeping conditions in Estonian milking cow’s udder diseases study, the 95% confidence limits for RR are approximately calculated as 95% CI RR e 1.94 1.96 1 190 1 4518 ;1.94 e1.96 1 190 1 4518 1.68; 2.24 . As this interval does not include 1, then there is less than a 1 in 20 chance that the reported difference between keeping conditions is solely due to chance. If the risk factor has more than two levels, then the relative risks can be calculated in relation to the different levels, usually the level of risk factor with the lowest incidence rate is used as the base. For example, the udder diseases incidences, incidence rates, relative risks and the confidence intervals of relative risks in case of different dung removal methods and in case of different types of bedding in Estonian milking cow’s study are presented in Table 3. Table 3. Udder diseases – incidence and relative risks by risk factors in Estonian dairy cows Risk factor Dung removal manual scaper tractor Type of bedding straw peat sawdust Number of observations Number of cases Incidence rate Relative risk 95% CI 848 29735 52041 12 1548 3148 0.014 0.050 0.057 1.00 3.54 4.07 2.01–6.25 2.31–7.17 14952 47879 19793 540 3156 1012 0.035 0.062 0.049 1.00 1.77 1.39 1.62–1.94 1.25–1.54 Other commonly used disease status measures in epidemiological studies are odds and odds ratios. The odds of an event are calculated as the number of events divided by the number of non-events. For example, on average 2000 drones are born in every 60000 births in beehive during year, so the odds of any randomly chosen bee being that of a drone is: number of drones number of queens 2000 58000 0.034 . Equivalently we could have calculated the same answer as the ratio of the bee being a drone (0.033) and it not being a drone (0.967). If the odds of an event are greater than one the event is more likely to happen than not; if the odds are less than one the chances are that the event won't happen. When events are rare, risks and odds are very similar. For example, in the bee’s sex example 2000 of 60000 born bees were drones: a risk of 0.033 [2000/60000] or an odds of 0.034 [2000/(60000-2000)]. Odds ratio (OR; synonyms: cross-product ratio, relative odds) is the probability of the event divided by the probability of the nonevent. It is a measure of the degree of association – for example, the odds of exposure among the cases (receiving the treatment of interest or exposed to a risk factor) compared with the odds of exposure among the controls (the risk factor or intervention is not present). For odds ratio the asymptotic confidence limits can be found using the following formula: 95% CIOR eln(OR)1.96se[ln(OR)] e OR 1.96se[ln(OR)] ;OR e1.96se[ln(OR)] , where se[ln(OR)] 1 number of cases in exposed animals 1 number of cases in non-exposed animals 1 number of controls in exposed animals 1 number of controls in non-exposed animals . When (disease) events are rare (which is usual in veterinary medicine), the estimates of RR are similar to those of OR. For example, comparing the keeping conditions in Estonian milking cow’s udder diseases study, the OR and its approximated 95% CI are calculated as OR 95% CIOR e 4518 190 0.0593 2.00 , (80732 4518) (6600 190) 0.0296 2.00 1.96 1 190 1 45181 6410 1 76214 ; 2.00 e1.96 1 190 1 45181 6410 1 76214 1.73; 2.32 . This suggests that those cows who are not kept in free stalls, are almost 2 times more likely diseased in udder diseases than those who are living in farms with tie keeping. Similarly to the relative risk, in case of risk factors with more than two levels, the odds ratios can be calculated in relation to the different levels. Odds ratios are the main parameters used in hypothesis testing and model building in epidemiological studies. Epidemiological studies generally try to identify factors that cause harm – those with odds ratios greater than one (contrary to clinical trials, where typically is looked for treatments which reduce event rates, and which have odds ratios of less than one). For example, the tie keeping can cause more udder diseases compared with free stall keeping. The “logit” model The "logit" model is used instead of standard regression and analysis of variance (ANOVA) models, if the dependent variable is binary and is measured on the 0/1-scale. For example the disease status (healthy/diseased), pregnancy status, treatment effect (no/yes). The logistic regression model has a form logit( ) ln[ (1 )] x , or (1 ) e x , where ln is the natural logarithm, e is the known constant (e = 2.71828… ); is the probability that the studied event occurs, for example, the animal is diseased; (1 ) is the odds ratio and ln[ (1 )] is the log odds ratio, or “logit”; x is the independent variable (argument); , and are respectively the regression coefficients and random error term, like in standard regression analyses. The logistic regression model is simply a non-linear transformation of the linear regression. The "logistic" distribution is an S-shaped distribution function, which constrains the estimated probabilities to lie between 0 and 1. From the logistic regression model the estimated probability is expressed as: x e x . 1 e Now it is evident, that if you let x 0 , then 0.5 ; as x gets really big, approaches 1; and as x gets really small, approaches 0. For example, assuming that all observations in Estonian milking cow’s study are independent, then the udder diseases incidence ( ) is predictable by stall length (SL) with simple logistic regression model of the form 7.546 0.028*SL e 7.5460.028*SL . 1 e The graphical representation of last model is visible on Figure 2. Figure 2. The udder diseases incidence in Estonian dairy cows predicted by stall length Mastitis incidence 0,06 0,05 π 0,04 e 0,591 0,015*X 1 e 0,591 0,015*X 0,03 0,02 0,01 140 160 180 200 220 240 Stall lenght The exponent of regression coefficient , e , is interpreted as the change in odds ratio corresponding to the one unit change in independent variable. For example, if e 2 , then a one unit change in independent variable would make the event twice as likely to occur. Negative regression coefficients lead to odds ratios less than one: if e 1 , then a one unit change in independent variable leads to the event being less likely to occur. For example, in Estonian milking cow’s study the regression coefficient allowing to predict the changes in udder disease incidences based on stall length, is 0.028. Thus the odds ratio corresponding to the 1 cm change in stall length is equal to e0.028 1.03 ; the odds ratio corresponding to the 10 cm change in stall length is equal to e0.028*10 1.32 . Usually the logistic regression analysis is performed with help of statistical analysis programs (SAS, R, SPSS, …) and the output contains additionally information about the exact confidence limits of estimated parameters and p-values corresponding to the tests of parameters statistical significance. If only confidence intervals to regression coefficients are printed out, then the confidence intervals to odds ratios can be calculated by applying the exponent function to coefficient CI’s. The part of standard output of SAS procedure LOGISTIC is presented on Figure 3. Figure 3. The part of standard output of SAS procedure LOGISTIC The LOGISTIC Procedure Analysis of Maximum Likelihood Estimates Parameter DF Estimate Standard Error Wald Chi-Square Pr > ChiSq Intercept SL 1 1 -7.5463 0.0279 0.2824 0.00167 713.9724 278.1635 <.0001 <.0001 Odds Ratio Estimates Effect SL Point Estimate 1.028 95% Wald Confidence Limits 1.025 1.032 Since usually the investigated diseases are multifactorial, i.e. many different factors participate in their aetiology, it is natural to analyse different risk factors in the context of a single complex model, also taking into consideration possible confounding influences. For this the generalized linear models with logistic link function can be used. For example, in Estonian milking cow’s udder diseases study, the complex model of the following form was used: logit() = + Ki + DRj + BTk + Fl + YMm + b1*SLijklmno + Ln + ijklmno, where is disease incidence, is the intercept, Ki is the influence of housing type i, DRj is the influence of manure removal j, BTk is the influence of type of bedding k, Fl is the influence of the farm l, YMm is the influence of year-month combination m, SLijklmno is stall length and b1 is the corresponding regression coefficient, Ln describes the effect of the repeated measurement of the nth cow, and ijklmno designates the portion of the value of the investigated attribute that failed to be described by the factors (random error). From such models, where the possible confounding influences are taken into the consideration, adjusted odds ratios (AOR, sometimes named also as adjusted relative risks) can be estimated by applying the exponent function to the assessments of the parameters of the model issued by the computer. The part of standard output of SAS procedure GENMOD, used to fit the abovementioned model with udder diseases data in Estonian milking cow’s study, is presented on Figure 4. From parameter estimates table in Figure 4 the adjusted odds ratios and their confidence intervals can be calculated and in last table the Wald Type 3 test results about factors significance are presented. Figure 4. The part of standard output of SAS procedure GENMOD Analysis Of GEE Parameter Estimates Empirical Standard Error Estimates Parameter Estimate Standard Error Intercept -6.6032 0.8732 95% Confidence Limits Z Pr > |Z| -8.3145 -4.8918 -7.56 <.0001 0.3711 0.0000 -2.1325 -1.7744 0.0000 1.1731 1.8845 0.0000 0.0006 0.9843 0.0000 -0.8214 -0.5673 0.0000 3.4647 4.2375 0.0000 0.0181 4.33 . -4.42 -3.80 . 3.97 5.10 . 2.09 <.0001 . <.0001 0.0001 . <.0001 <.0001 . 0.0366 …… KEEP KEEP BE_TYPE BE_TYPE BE_TYPE DU_REM DU_REM DU_REM ST_LENGTH 1 3 1 2 3 1 2 3 0.6777 0.0000 -1.4770 -1.1709 0.0000 2.3189 3.0610 0.0000 0.0094 0.1564 0.0000 0.3345 0.3079 0.0000 0.5846 0.6003 0.0000 0.0045 Wald Statistics For Type 3 GEE Analysis Source DF ChiSquare Pr > ChiSq KEEP BE_TYPE DU_REM FARM YEAR_MON ST_LENGTH 1 2 1 10 38 1 18.77 19.50 93.52 504.74 253.67 4.37 <.0001 <.0001 <.0001 <.0001 <.0001 0.0366