The Microbiology of Bad Breath and Periodontitis

The Microbiology of Bad
Breath and Periodontitis
By Kate Egge
1
Overview
• Background information about bad breath and
Periodontitis
• Discuss 3 bacteria that cause oral malodor
• Methods for detecting bacteria and?or Bad breath:
PCR, organoleptic, VSC concentrations, and BANA tests
• Research: that indicates bacteria cause bad breath
• Research: that indicates that bacteria may cause periodontitis
• Conclusions
• References
• Questions
2
Bad Breath
• Americans spent 625 million dollars on breath fresheners in
2000.
• 85-95% of cases originate in the mouth
• Microorganisms on the posterior dorsal side of tongue is the main source of bad breath
• Area is poorly cleansed by saliva and contains invaginations where bacteria can hide and putrefy postnasal drip
3
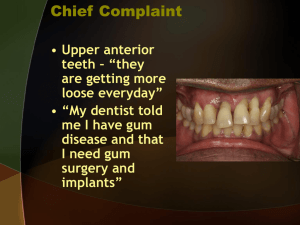
Periodontitis
• Some bad breath is associated with periodontal disease and destruction of gums
• Periodontitis is a common, progressive disease that affects the supporting structures of the teeth, causing loss of attachment to the bone and often resulting in tooth loss
4
Bad breath
• Bad breath primarily result of microbial metabolism.
• Organisms feed off of glycoproteins and release offensive chemical odors
• Oral gram-negative organisms such as spirochetes and some bacteroides have the capacity to produce oral malodor through putrification of sulfur containing protein substrates such as cysteine and methionine
• Chemistry
5
Important microorganisms
• Key anaerobic organisms
Treponema denticola
,
Porphyromonas gingivalis,
and
Bacteroides forsythus
6
Phylogenetic tree of major lineages of Bacteria based on 16s ribosomal
RNA sequence comparisons
7
Treptonema denticola
• Gram negative, anaerobic spirochete
• A lot of size variation among the spirochetes
• Form a major phylogenetic lineage of Bacteria
• Protoplasmic cylinder
• Flagella emerge from each pole and are located in the periplasm of the cell (endoflagella)
• Endoflagella and protoplasmic cylinder are surrounded by a flexible membrane called the outer sheath
8
Porphyromonas gingivalis
and
Bacteroides forsythus
• Member of genus Bacteroides
• Gram negative obligate anaerobe
• Ferments sugars to primarily acetate and succinate
• Bacteroides species dominant bacteria in large intestine, found 10^10 – 10^11 cells per gram of human feces
9
Porphyromonas gingivalis
• Several clinical studies show that
P. gingivalis
is a factor in periodontitis
• Study Journal of Clinical Microbiology compared the prevalence of
P. gingivalis
a group with periodontitis to a different group that was periodontally healthy in
• Results:
10
Porphyromonas gingivalis
•
P. gingivalis
was detected in only 25% of the healthy subjects but was detected in
79% of the periodontitis group
• The odds ratio for being infected with
P. gingivalis
was 11.2 times greater in the periodontits group than in the healthy group.
11
Porphyromonas gingivalis
and
PCR
• PCR assays were used for detection of
P.gingivalis
• Target sequence was the ribosomal DNA spacer region between 16S and 23S ribosomal genes
• DNA fragments were separated by agarose gel electrophoresis
12
Detecting
Porphyromonas gingivalis
13
Methods for detecting oral malodor
• Organoleptic measurements:
• Scale 0=no odor up to 5=foul odor
• Tongue malodor was assessed by scraping the posterior tongue dorsum with gauze and immediately smelling it’s odor.
• Malodor in dental sites was assessed by pushing waxed dental floss between teeth and immediately smelling and recording the odor based on the organoleptic scale
14
Methods for detecting oral malodor continued…
• High VSC concentrations indicate oral malodor
• Volatile Sulfur Compounds (VSC) are measured with a portable sulfide monitor
• VSC monitor contains a pump which sucks air from a plastic straw placed inside the subjects mouth
• The mouth air is passed through an electrolytic sensor which measures the concentration of volatile sulfurs
• VSC levels are strongly associated with organoleptic scores
15
Methods for detecting oral malodor continued
• BANA test
Test kit for bacterial trypsin like activity based on the hydrolysis of benzoyl-DL-arginine-naphthylamide (BANA) used to asses the presence of one or more of
T. denticola , P. gingivalis , B. forsythus
• These bacteria are capable of hydrolyzing the synthetic trypsin substrate BANA- an activity not identified in other oral microbial species
• Conditions favoring a Positive BANA test on the tongue dorsum and tooth sites were associated with oral malodor production
16
Relationship of Oral Malodor to
Periodontitis
• Species identified by BANA tests have been tentatively implicated as putative periodontal pathogens
• However, some studies have found that species
Treponema denticola , Porphyromonas gingivalis, and Bacteroides forsythus identified by BANA tests do not cause periodontitis.
• When peridontitis was present 87.5% of tooth sites and 92.5% of tongue sites were BANA (+)
• In healthy individuals 74.4% of tooth sites and
93% of tongue sites were BANA (+)
17
Relationship of Oral Malodor to
Periodontitis continued
• Oral malodor is not directly associated with presence of periodontitis
• Substantial oral malodor may be present in individuals without periodontitis
18
Conclusions
• Gram negative anaerobic oral bacteria ( Treponema denticola ,
Porphyromonas gingivalis, and Bacteroides forsythus ) cause bad breath
• Key anaerobic organisms Treponema denticola , Porphyromonas gingivalis, and Bacteroides forsythus degrade glycoproteins proteins and release foul smelling chemicals
• Strong evidence indicates that these bacteria cause bad breath
• Treponema denticola: Anaerobic spirochete, Protoplasmic cylinder and endoflagella
• Porphyromonas gingivalis, and Bacteroides forsythus are both m embers of the genus Bacteroides
19
Conclusions continued
• Bacteria on the posterior dorsal side of tongue is the main source of bad breath
• One study using PCR found that P.gingivalis was 11x more likely to be found in individuals that had periodontitis- P. gingivalis may be a periodontal pathogen
• However, research using BANA tests found that the presence of T. denticola , P. gingivalis, and B. forsythus was NOT associated with periodontitis
• Oral malodor is not directly associated with the presence of periodontitis
20
References:
• Rosenberg, Mel. Clinical Assessment of Bad Breath: Current Concepts.
Journal of the American Dental Association 1996; 127: 475-82
• Griffen A, Becker M, Lyons S. Prevalence of Porphyromonas gingivalis and
Periodontal Health Status. Journal of Clinical Microbiology 1998; 36(11)
:3239-42
• Madigan M, Martinko J, Parker J. Biology of Microorganisms. Ninth edition
• A, Bosy, G.V. Kulkarni, M. Rosenberg and C.A.G. McCulloch. Relationship of oral malodor to periodontitis: Evidence of independence in discrete subpopulations. Journal of Periodontology 1994; 65:37-46.
• Rosenberg Mel. The science of Bad Breath. Scientific American 2002; 72-79.
• Bruce P,Boches S, Galvin J. Bacterial Diversity in human subgingival plaque.
Journal of Bacteriology 2001;183(12): 3770-83.
• De Boever E, Uzeda M, Loesche W. Relationship between volatile sulfur compounds,Bana-Hydrolyzing bacteria and gingival health in patients with and without complaints of oral malodor. Journal of Clinical Dentistry 1994;
4(4):114-119
21
22
23
24
25
26