A Survey of the Microbial World Chapter 10
advertisement

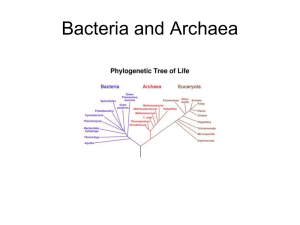
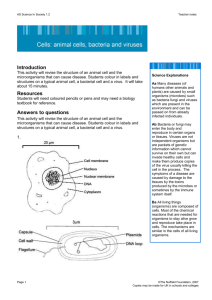
A Survey of the Microbial World Chapter 10- Classification of Microorganisms Siti Sarah Jumali Room 14 Level 3 (ext 2123) www.slideshare.net/sarah_jumali The prokaryotes kingdom • The prokaryotes kingdom – Bacteria and Archaea – Classification and identification – Bergey’s manual and bacteria taxonomy – Biochemical tests Evolution • Similarities among organisms such as having plasma membrane, the use of ATP, and possessing DNA are the result of evolutiondescent from common ancestor • Darwin (1859) says natural selection was responsible for similarities and difference among organisms. Evolution cont’d Phylogenetic relationship Evolution can be deduced from phylogeny. • Taxa (singular: taxon)- a group of one or more organisms • Taxonomy- put organisms into categories (taxa) to show degrees of similarities between organisms • Phylogeny or systematics- study of evolutionary history of organisms The Domain of Life (The 3 Domains) • Comparing cells through ribosomes – Ribosomes are different in every cells – Ribosomes are present in all cells – Comparing the sequences in ribosomal RNA (rRNA) gives 3 distinct groups: • Eukaryotes • Prokaryotes-bacteria and archaea (2 different types or prokaryotes) • Differ in membrane lipid structure, transfer RNA molecules (tRNA) and sensitivity to antibiotics Eukarya • • • • Animals Fungi Plants Protists Eukarya Prokayotes and Archaea • Prokaryotes – pathogenic and non-pathogenic – Found in soil and water • Archaea – includes prokaryotes that do not have peptidoglycan in their cell walls – Live in extreme environments and carry out unusual metabolic processes – Includes • Methanogens ( strict anaerobes that produce CH4 from CO2 and H2 • extreme halophiles • hyperthermophiles The Domain of Life (The three Domains) Origin of mitochondria Origin of choroplast DNA passed from ancestors are called conserved Nucleoplasm grows larger Endosymbionts • Endosymbiotic theory – Eukaryotic cells evolved from prokaryotic cells – Eukaryotic cells and prokaryotic cells live in one another (mitochondria and chloroplast) The 3 Domains And..only bacteria is sensitive to antibiotics Classification of microorganisms • Scientific nomenclature – Genus – Specific epithet (species) Binomial nomenclature – Eg. Rhizopus stolonifer –rhizo –rootlike structure of fungus, - stolo- long hyphae – Enables identification of fungus which tells us the right treatment that can be used – Follows the trend • Genus, Family, Order, Class, Phylum, Kingdom, Domain Classification of Prokaryotes • Found in Bergey’s manual of systematic bacteriology • Prokayotes are divided into 2 domains- abcteria and archaea • Domain divided into phyla • Based on rRNA sequence similarities • Class Order Family Genera Species Classification of Prokaryotes cont’d • Eukaryotes- a group of closely related organisms that can interbreed • Prokaryotes- cell division is indirectly tied to sexual conjugation, infrequent and not necessarily species-specific – Therefore termed as population of cells with similar characteristics – Bacteria grown in media at a given time are culture – Pure culture is often a clone – But in some cases, the same species are dissimilar in all ways, therefore called a strain Classification of Eukaryotes • • • • Animals Fungi Plants Protists Eukaryotic Cells Classification of Viruses • Not part of either domains • Not composed of cells • Use anabolic machinery within host cell to multiply • Viral genome can direct biosynthesis inside a cell • Some can be incorporated into the host’s genome • Virus is more closely related to its host than to other virus • Viral species- morphology, genes, enzymes • Obligatory intracellular parasites Viruses • Hypotheses on the origin of viruses 1) Arose independently replicating strands of nucleic acids such as plasmids 2) They developed from degenerative cells that through many generations they gradually lost the ability to survive independently but could survive when associated with another cell Virus Methods of Classifying and Identifying organisms • Classification – Identification is for practical purposes • E.g to determine appropriate treatment of infection – Can be identified microscopically • Morphology – Bergey’s manual of Determinative Bacteriology – Based on criteria • • • • • Cell wall composition, morphology, differential staining, oxygen requirements and; biochemical test Methods of Classifying and Identifying organisms cont’d • Source and habitat of isolate are a part of identification • In clinical microbiology, • Information returned will allow further treatment • uses transport media (from swab) – Not nutritive – Transport media prolongs viability of fastidious organisms Identification of microorganisms • Morphological Characteristics – Cocci, rod, spirilla – Tells us little about phylogenetic relationship – Differences in structures such as endospores and flagella • Differential staining – Gram stain and acid-fast stain – Based on chemical composition of cell wall – Not useful for identification of wall-less and archaea with unusual cell wall Identification of microorganisms Biochemical tests • Ability to ferment certain type of carbohydrate • Gram identification – All members of the family Enterobacteriacaea are oxidase-negative • Genera Escherichia, Enterobacter, Shigella, Citrobacter and Salmonella • All of these ferment lactose to produce acid and gas except for Shigella and Salmonella • Selective media • Differential media • Rapid identification Biochemical tests Biochemical tests Rapid Identification Method • Manufactured for medically important group such as enterics • Perform biochemical tests simultaneously • Identify bacteria within 4-24 hours • Numerical identification- result is assigned with number – eg positive = 1, negative = 0 – Results are compared to a database of unknown organisms Rapid Identification Method cont’d Rapid Identification of bacteria using Becton Dickinson Serology • Study of serum and immune responses • Microorganisms are antigenic, they stimulate antibodies • Eg inject a rabbit with killed typhoid bacteria and the rabbit will produce antibody agaisnt typhoid bacteria • Solution of antibody-Antiserum • Slide agglutination test– put sample on slide and add different known antiserum – Bacteria will agglutinate when mixed with antibodies in response to species or strain of bacterium – Positive test is observed in the presence of agglutination Serology cont’d • Serological testing – Can differentiate not only among microbial species but also strains within species – Also include ELISA and Western Blotting • Strains with different antigens are called serotypes, serovars or biovars – E.g Different antigens in the cell walls of various serotypes of streptococci stimulate different antibodies Serology cont’d • But because closely related bacteria also produce some similar antigens, – serological testing can be used to screen bacteriological isolates for possible similarities • ELISA (Enzyme linked Immunosorbent Assay) – Used to detect AIDS – Fast and can be read by computer scanner – Known antibodies are placed in microplate wells, and bacteria are added into it – Reaction produced allows identification of bacteria Serology cont’d Western blot Western blot ELISA ELISA cont’d Other techniques for identification • • • • • • • Phage typing Fatty acids profile Flow cytometry DNA base composition DNA fingerprinting Polymerase Chained Reaction (PCR) Nucleic Acid Hybridization Other techniques for identification • Phage typing – Looks for similarities among bacteria – Useful to trace the origin and the cause of outbreak – determines which phage the bacterium is susceptible to • Fatty acids profile – Fatty Acids methyl esther (FAME) – Separate cellular fatty acids and compare to fatty acids profile of known organisms Flow cytometry – Identify bacteria without culturing – Detect difference in electrical conductivity between cells and the surrounding medium – Provide information on the size, shape, density and – Can detect fluorescent cells such as Pseudomonas or fluorescent tagged cells Flow cytometer Other techniques for identification • DNA base composition – Percentage of guanine and cytosine (G + C), indirectly tells the composition of (A + T) – Similar organisms may have about the same GC percentage • DNA fingerprinting – RFLP is used to break DNA fragments (Restriction enzymes) – Determine source of hospital-acquired infection • Polymerase Chained Reaction (PCR) – Increase amount of DNA – Detects presence of microorganisms on gel electrophoresis – Taq polymerase Nucleic acid Hybridization • Southern Blotting – The use of probe • DNA chips – The use of probe – Fluorescent dye • Ribotyping and rRNA sequencing – All cells contain RNA – RNA underwent lots of changes over time – Does not require cells to be cultured in the laboratory • FISH (Fluorescent In Situ Hybridization) – Fluorescent-dye labeled – Can be used to detect bacteria in drinking water or in patient less than 24 hr Nucleic acid Hybridization FISH- Ikan Putting Classification Methods Together • Dichotomous keys – Identification based on successive questions: Yes/ No, Positive/ Negative • Cladograms – Branches – Using computer Questions???