Negative Binomial Regression - NASCAR Lead Changes (1975
advertisement
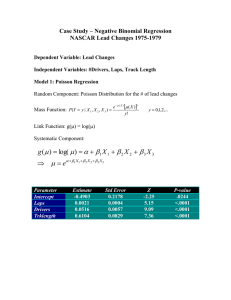
Negative Binomial Regression NASCAR Lead Changes 1975-1979 Data Description • Units – 151 NASCAR races during the 19751979 Seasons • Response - # of Lead Changes in a Race • Predictors: # Laps in the Race # Drivers in the Race Track Length (Circumference, in miles) Models: Poisson (assumes E(Y) = V(Y)) Negative Binomial (Allows for V(Y) > E(Y)) Poisson Regression • Random Component: Poisson Distribution for # of Lead Changes • Systematic Component: Linear function with Predictors: Laps, Drivers, Trklength • Link Function: log: g(m) = ln(m) y m X m X e Mass Function: P Y y | X 1 , X 2 , X 3 y! g m X 1 X 1 2 X 2 3 X 3 x ' m X e 1 X1 2 X 2 3 X 3 e x ' x ' 1 X 1 X 2 X 3 Regression Coefficients – Z-tests Parameter Intercept Laps Drivers Trklength Estimate -0.4903 0.0021 0.0516 0.6104 Std Error 0.2178 0.0004 0.0057 0.0829 Z -2.25 5.15 9.09 7.36 Note: All predictors are highly significant. Holding all other factors constant: • As # of laps increases, lead changes increase • As # of drivers increases, lead changes increase • As Track Length increases, lead changes increase m e 0.49030.0021L0.0516 D0.6104T P-value .0244 <.0001 <.0001 <.0001 Testing Goodness-of-Fit • Break races down into 10 groups of approximately equal size based on their fitted values • The Pearson residuals are obtained by computing: ^ ^ Yi m i Yi m i observed- fitted ei ^ V (Yi ) fitted mi X 2 ei2 • Under the hypothesis that the model is adequate, X2 is approximately chi-square with 10-4=6 degrees of freedom (10 cells, 4 estimated parameters). • The critical value for an =0.05 level test is 12.59. • The data (next slide) clearly are not consistent with the model. • Note that the variances within each group are orders of magnitude larger than the mean. Testing Goodness-of-Fit Range 0-9.4 9.4-10.5 10.5-11.6 11.6-20 20-21 21-23 23-26 26-32 32-36 36+ Total #Races 15 14 14 17 19 15 16 16 11 13 151 obs fit 113 138 178 321 485 191 353 491 349 574 131.3 150.6 157.1 274.4 390.3 328.7 397.1 452.9 374.2 536.4 Pearson -1.60 -1.03 1.67 2.81 4.79 -7.60 -2.21 1.79 -1.30 1.62 2 X =107.4 Mean 7.53 9.20 12.71 18.88 25.53 12.73 22.06 30.69 31.73 44.15 Variance 23.41 34.46 41.30 56.36 89.93 48.21 74.33 183.70 201.82 229.47 107.4 >> 12.59 Data are not consistent with Poisson model Negative Binomial Regression • Random Component: Negative Binomial Distribution for # of Lead Changes • Systematic Component: Linear function with Predictors: Laps, Drivers, Trklength • Link Function: log: g(m) = ln(m) y k k k m Mass Function: P Y y | X 1 , X 2 , X 3 , k k y 1 k m k m E Y m V Y m m2 k g m X 1 X 1 2 X 2 3 X 3 x ' m X e 1 X1 2 X 2 3 X 3 e x ' x ' 1 X 1 X 2 X 3 y y 0,1, 2,... Regression Coefficients – Z-tests Note that SAS and STATA estimate 1/k in this model. Parameter Intercept Laps Drivers Trklength 1/k m e Estimate -0.5038 0.0017 0.0597 0.5153 0.1905 Std Error 0.4616 0.0009 0.0143 0.1636 0.0294 Z -1.09 2.01 4.17 2.87 0.5038 0.0017 L 0.0597 D 0.5153T V Y m 0.1905 m 2 P-value .2752 .0447 <.0001 .0041 Goodness-of-Fit Test Pearson Residuals: ei Range 0-9.4 9.4-10.5 10.5-11.6 11.6-20 20-21 21-23 23-26 26-32 32-36 36+ Total #Races 13 17 20 11 21 12 18 16 14 9 151 Yi m i V Yi obs fit 96 155 248 251 523 141 442 470 445 422 111.4 170.2 223.3 202.4 431.5 261.8 452.0 464.3 485.3 397.5 Yi m i 2 mi mi k Pearson -0.30952 -0.20153 0.250504 0.543148 0.482911 -1.04674 -0.0504 0.02797 -0.18924 0.140292 X2=1.88 X 2 ei2 Mean 7.38 9.12 12.40 22.82 24.90 11.75 24.56 29.38 31.79 46.89 S.D. 4.22 6.10 5.87 5.53 9.55 6.44 10.98 14.83 14.12 13.82 • Clearly this model fits better than Poisson Regression Model. • For the negative binomial model, SD/mean is estimated to be 0.43 = sqrt(1/k). • For these 10 cells, ratios range from 0.24 to 0.67, consistent with that value. Computational Aspects - I k is restricted to be positive, so we estimate k* = log(k) which can take on any value. Note that software packages estimating 1/k are estimating –k* Likelihood Function: k yi k ( yi k ) k mi ( yi k 1) (k )(k ) k mi Li (k )( yi 1) k mi k mi (k )( yi 1) k m k m i i ( y k 1) i yi ! k yi ( k ) k mi k m k m i i ( yi e 1) yi ! k* ek* yi mi e e k* k* e m e m i i k* k* yi Log-Likelihood Function: yi 1 li ln Li ln(ek* j ) ln yi ! ek* ln(ek* ) yi ln( mi ) (ek* yi ) ln( mi ek* ) j 0 Computational Aspects - II Derivatives wrt k* and : yi 1 li e k * yi 1 k* k* e k * 1 ln(e ) k * ln(e k * mi ) k * e mi j 0 e j yi 1 yi 1 2li e k * yi mi yi 1 1 ek* k* k* k* k* k* e k * 1 ln(e ) k * ln(e mi ) e k * 1 e 2 k* 2 (k *) 2 e j e m ( e j ) m ek* j 0 i i m e j 0 i 2li y m k* i i xi e mi m e k * 2 k * i y m li xi e k * i ki* mi e k* 2li e yi k* xi xi ' e mi m e k * 2 ' i Computational Aspects - III Newton-Raphson Algorithm Steps: l gk i k * 2li Gk k *2 l g i 2li G ' g g k gk G k G ' 2 l i k * Step 1: Set k*=0 (k=1) and iterate to obtain estimate of : Step 2: Set ’ = [1 0 0 0] and iterate to obtain estimate of k*: 2li k * Gk * ~ (i ) ~ (i ) k* ~ ( i 1) ~ ( i 1) k* G ~ ( i 1) Gk 1 g k k * Step 4: Back-transform k* to get estimate of k: k=exp(k*) g Gk g k Step 3: Use results from steps and 2 as starting values (software packages seem to use different intercept) to obtain estimates of k* and ~ (i ) 1 1