Stacks - Weizmann Institute of Science
advertisement

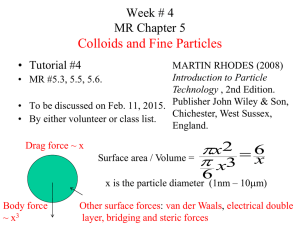
Digital Image Processing in Life Sciences April 18th, 2012 Lecture number 5: Image stacks- beyond 2D Today’s topics: Stacks: projections, 3D Kalman Filtering Tracking- spatial (morphology) temporal (trajectories) imageJ macros- if time permits Stacks “Multiple spatially or temporally related images in a single window”. Why do we acquire stacks? Temporal information- acquisition of time-lapse movies Spatial information- acquisition of Z-stacks Components information- acquisition at different wavelengths What do we do with stacks? Display our data- projections vs 3D renderings Analyze our data- construct tracks that represent movement and/or structures Typical analyses we perform on stacks: Particle movement-velocity, direction etc. Morphologies that extend into the Z dimension Structures that change their shape with time Stacks “Multiple spatially or temporally related images in a single window”. The images that make up a stack are called slices. 3D vs 4D vs 5D stacks: Spatially=z; temporally=t; channels=c. Combinations of xy with z/t/c. In stacks, a pixel becomes a voxel (volumetric pixel), i.e., an intensity value on a regular grid in a three dimensional space. Pixel X volumetric pixel=voxel y Z or t or C Stacks All the slices in a stack must eventually be the same size and bit depthBut not necessarily originally: Some softwares allow formation of stacks from slices that have different parameters/dimensions. Some commands found in imageJ, but not only: Hands on… “Rat_Hippocampal_Neuron”: [Stack To Images] [Images To Stack] [Make Montage] “Fly Brain”: [Reslice. . .] [Orthogonal Views] [Z project]: (average, max, min, std, sum, median) –one dimension is lost! [3D project] “Each frame in the animation sequence is the result of projecting from a different viewing angle. To visualize this, imagine a field of parallel rays passing through a volume containing one or more solid objects and striking a screen oriented normal to the directions of the rays. Each ray projects a value onto the screen, or projection plane, based on the values of points along its path. Three methods are available for calculating the projections onto this plane: nearest-point, brightestpoint, and mean-value. The choice of projection method and the settings of various visualization parameters determine how both surface and interior structures will appear.” (imageJ manual) Nearest Point projection - produces an image of the surfaces visible from the current viewing angle. Brightest Point projection- examines points along the rays, projecting the brightest point encountered along each ray. Mean Value projection- produces images with softer edges and lower contrast, but can be useful when attempting to visualize objects contained within a structure of greater brightness (e.g. a skull). Lower / Upper Transparency Bound Determines the transparency of structures in the volume. Projection calculations disregard points having values less than the lower threshold or greater than the upper threshold. Opacity Can be used to reveal hidden spatial relationships, specially on overlapping objects of different colors and dimensions. This can give the observer the ability to view inner structures through translucent outer surfaces. Surface / Interior Depth–Cueing Depth cues can contribute to the three-dimensional quality of projection images by giving perspective to projected structures. The depthcueing parameters determine whether projected points originating near the viewer appear brighter, while points further away are dimmed linearly with distance. The trade-off for this increased realism is that data points shown in a depth-cued image no longer possess accurate densitometric values. Why use 3D? Aren’t projections good enough? Maximal intensity projections can be misleading (3 dimensions are better than 2): Case 1- inferring wrong morphology Case 2- inferring wrong co-localization The Kalman Filter “Operates recursively on streams of noisy input data to produce a statistically optimal estimate of the underlying system state. It uses a series of measurements observed over time, containing noise and inaccuracies, and produces estimates of unknown variables that tend to be more precise than those that would be based on a single measurement alone.” “An Introduction to the Kalman Filter”, 2006, Welch and Bishop After each time and measurement update pair, the process is repeated with the previous a posteriori estimates used to project or predict the new a priori estimates. Prediction step: the Kalman filter estimates current state variables with their uncertainties. When the outcome of the next measurement is observed, these estimates are updated using a weighted average, with more weight being given to estimates with higher certainty. This way, values with smaller estimated uncertainties are "trusted" more. The weights are calculated from the covariance, which is a measure of the estimated uncertainty of the prediction of the system's state. The result of the weighted average is a new state estimate whose value is in between the predicted and the measured state, and has a better estimated uncertainty than either of them alone. This process is repeated every time step, with the new estimate and its covariance informing the prediction used in the following iteration. This means that the Kalman filter works recursively and requires only the last "best guess" - not the entire history - of a system's state to calculate a new state. The Kalman gain - a function of the relative certainty of the measurements and current state estimate. It can be adjusted to achieve desired performance: High gain: the filter places more weight on the measurements, and adheres to them more. Low gain: the filter follows the model predictions more closely, smoothing out noise but decreasing the responsiveness. At the extremes, a gain of one causes the filter to ignore the state estimate entirely, while a gain of zero causes the measurements to be ignored. “Methods for Cell and Particle Tracking” Erik Meijering, Oleh Dzyubachyk, Ihor Smal (2012) Generally two sides to the tracking problem: 1) the recognition of relevant objects and their separation from the background in every frame (the segmentation step), and 2) the association of segmented objects from frame to frame and making connections (the linking step). The objects are most easily segmented by thresholding, which labels pixels above the intensity threshold as “object” and the remainder as “background”, after which disconnected regions can be automatically labeled as different objects. In the case of severe noise, auto-fluorescence, photobleaching, poor contrast, gradients, or halo artifacts, thresholding will fail, and more sophisticated segmentation approaches are needed: •Template matching (which fits predetermined patches or models to the image data but is robust only if cells have very similar shape). •Watershed transformation (which completely separates images into regions and delimiting contours but may easily lead to over-segmentation). •Deformable models (which exploit both image information and prior shape information). The simplest approach to solving the subsequent association problem is to link every segmented cell in any given frame to the nearest cell in the next frame, where “nearest” may refer to spatial distance but also to difference in intensity, volume, orientation, and other features. Template matching, mean-shift processing, or deformable model fitting can be applied to one frame, and the found positions or contours are used to initialize the segmentation process in the next frame, and so on, which implicitly solves the linking problem. An example of a software for Morphological Tracing: Simple Neurite Tracer http://fiji.sc/wiki/index.php/Simple_Neurite_Tracer “Easy semi-automatic tracing of neurons or other tube-like structures (e.g. blood vessels)” •A filtering process that searches for geometrical structures which can be regarded as tubular. •A probe kernel that measures the contrast between the regions inside and outside the range. •Curvatures are then estimated. •Paths are found via a bidirectional search An example of a software (Matlab-based) for Single Particle Tracking: “Robust single-particle tracking in live-cell time-lapse sequences” Jaqaman et al., Nat. Meth., 2008. SPT- Establishment of correspondence between particle images in a sequence of frames. This is complicated by various factors: high particle density, particle motion heterogeneity, temporary particle disappearance, particle merging and particle splitting. The algorithm first links the detected particles between consecutive frames, and then links the track segments generated in the first step to simultaneously close gaps and capture particle merge and split events. The latter step ensures temporally global optimization. First step (particle assignment): link detected particles between consecutive frames. The condition: a particle in one frame could link to at most one particle in the previous or the following frame. The track segments obtained in this step tended to be incomplete, resulting in a systematic underestimation of particle lifetimes. The one-to-one assignment excludes splits and merges. Second step (track assignment): link initial track segments in three ways: (i) end to start, to close gaps resulting from temporary disappearance, (ii) end to middle, to capture merging events, and (iii) start to middle, to capture splitting events. Every potential assignment is characterized by a cost C. The goal of solving in each step is to identify the combination of assignments with the minimal sum of costs. Jaqaman et al., Nat. Meth., 2008 The intensity factor increased the cost when the intensity after merging or before splitting was different from the sum of intensities before merging or after splitting, with a higher penalty when the intensity was smaller. This intensity penalty ensured that merging and splitting events were not picked up only because of the proximity of particle tracks but that the associated intensity changes were consistent with the image superposition of merging or splitting particles. Jaqaman et al., Nat. Meth., 2008 Jaqaman et al., Nat. Meth., 2008 An example of a software (imageJ plugin) for Speckle Tracking: http://athena.physics.lehigh.edu/speckletrackerj/ Neuron_Morpho: http://www.personal.soton.ac.uk/dales/morpho/ NeuronJ http://www.imagescience.org/meijering/software/neuronj/ (Handles only two-dimensional (2D) images of type 8-bit gray-scale or indexed color) Jfilament (has 3D option) http://athena.physics.lehigh.edu/jfilament/ Danuser lab (including micro-track): http://lccb.hms.harvard.edu/software.html Fluorender http://www.sci.utah.edu/software/13-software/127-fluorender.html TrakEM2 in FIJI Neuromantic http://www.reading.ac.uk/neuromantic/ Kalman: http://rsbweb.nih.gov/ij/plugins/kalman.html Do not forget to acknowledge use of plugins developed by the community. Specific citation instructions are usually found on their websites. END!