B. Honarparvar, H.G. Kruger, T. Govender, GEM. Maguire
advertisement

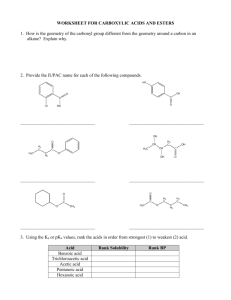
Pentacycloundecane lactam vs lactone norstatine type HIV protease inhibitors: binding energy calculations and DFT study B.Honarparvar, H.G. Kruger, T. Govender, G.E. M. Maguire Catalysis and Peptide Research Unit, School of Health Sciences, University of KwaZulu-Natal, South Africa Honarparvar@ukzn.ac.za December 2013 Structures of PCU-lactam-EAIS, its tautomer PCU-lactimEAIS, PCU-lactone-EAIS inhibitors and PCU-models OH OH O O HN OH NH O OH N O NH O O OH NH HN NH HN H2 N H2 N HN O O HN O O O OH OH PCU-Lactam-NH-EAIS PCU-Lactim-NH-EAIS IC50= 0.076 µM OH O O OH NH H N O O OH H 2N O O NH HN O O PCU-Lactone-CO-EAIS IC50= 0.850 µM OH OH O O O O OH OH NH NH HN O H 2N H N O NH HN O OH O O O O PCU-Lactim-CO-EAIS PCU-Lactam-CO-EAIS OH O HN OH O O O NH O O O NH HN H2 N OH PCU-Lactone-NH-EAIS HN H3 C HN OH H3 C HN O N OH NH HN O OH H 2N O H N O OH N O OH PCU-lactim PCU-lactam O OH H 3 C NH O O PCU-lactone Objectives MD simulation Binding free energy calculations DFT study Binding free energy calculations Software: Amber12 Method: MMPB(GB)SA PCU- peptide inhibitor inside the active site of South African HIV protease PCU-peptide inhibitor docked to HIV protease MM-PB(GB)SA binding free energy calculations The MM-PB(GB)SA method can be conceptually summarized as: ΔGbind = Gcomplex – (Genzyme + Gligand) where Gcomplex, Genzyme and Gligand are the free energies of the complex, the enzyme and the ligand, respectively. Binding free energies and its components for the PCU-models complexed with the HIV protease HN H3C OH N O HN H3C OH HN O OH PCU-lactim O OH H3C NH O PCU-lactam O O PCU-lactone ΔESOL( ΔESOL(G ΔGbind(PB ΔGbind(GBS PB) B) SA) A) ΔEELE ΔEVDW PCU-lactam(a) -1.48 -34.13 28.73 11.34 -29.52 -27.63 PCU-lactam(b) -1.88 -30.14 22.44 14.99 -29.69 -19.99 PCU-lactim(a) -12.23 -30.14 33.92 21.01 -29.85 -24.78 PCU-lactim(b) -9.52 -31.07 33.63 22.76 -27.85 -20.98 PCU-lactone(a) -2.51 -25.61 16.95 13.34 -28.19 -17.37 PCU-lactone(b) -5.30 -27.26 18.40 17.53 -23.39 -17.92 PCU-models Binding free energies and its components for the PCU-peptides complexed with the HIV protease OH OH O HN OH NH O NH O NH HN H2N O OH HN O O O O O H 2N OH PCU-Lactam-NH-EAIS NH HN O OH O O O PCU-Lactone-CO-EAIS IC50= 0.076 µM PCU-peptides H N O IC50= 0.850 µM ΔEELE ΔEVDW ΔESOL( ΔESOL(G ΔGbind(PB ΔGbind(G PB) B) SA) BSA) PCU-lactam-NH-EAIS(a) -29.39 -68.41 52.81 53.46 -69.79 -61.74 PCU-lactam-NH-EAIS(b) -19.65 -60.67 49.97 41.95 -64.39 -43.19 PCU-lactim-NH-EAIS(a) -30.39 -71.48 80.90 55.23 -68.07 -54.33 PCU-lactim-NH-EAIS(b) -21.94 -68.36 62.31 43.20 -72.54 -54.48 PCU-lactone-CO-EAIS(a) -24.22 -55.02 93.73 87.38 -61.77 -37.99 -29.39 -68.41 52.81 53.46 -69.79 -61.74 PCU-lactone-CO-EAIS(b) Binding free energies and its components for the synthesized PCU-peptides complexed with the HIV protease OH O OH O O OH NH HN O H2N H N NH HN HN O NH O O OH O O O O O NH HN O O OH H2N PCU-Lactam-CO-EAIS OH PCU-Lactone-NH-EAIS PCU-peptides ΔEELE ΔEVDW PCU-lactam-CO-EAIS(a) -23.21 PCU-lactam-CO-EAIS(b) ΔESOL( ΔESOL(G ΔGbind(P ΔGbind( PB) B) BSA) GBSA) -74. 82 117.34 106.88 -87.89 -58.79 -28.45 -69.76 123.84 99.49 -61.99 -40.02 PCU-lactim-CO-EAIS(a) -11.87 -59.99 56.69 37.49 -55.37 -55.99 PCU-lactim-CO-EAIS(b) -21.94 -68.36 62.31 43.20 -54.77 -72.54 PCU-lactone-NH-EAIS(a) -19.50 -74.31 73.49 46.27 -67.41 -55.34 -9.01 -74.08 52.55 34.55 -78.71 -57.10 PCU-lactone-NH-EAIS(b) DFT study of PCU-models Software: Gaussian09 Method: B3LYP Basis set: 6-311G** HN H3C OH N O OH PCU-lactim HN H3C OH HN O O PCU-lactam O OH H3C NH O O PCU-lactone Electronic structure properties Polarizability NBO Analysis (HOMO- LUMO) Natural atomic charges Electrostatic Potential Map Natural atomic charges (a.u.) on nitrogen and oxygen nuclei of PCU-models 4 HN O 5 HN 1 OH 3 O 5 O 2 PCU-lactam 5 O 4 HN N 1 OH 3 OH 2 PCU-lactim NH 4 O 1 OH 3 O 2 PCU-lactone Atom Lactam Lactim Lactone N1/O1 -0.65213 -0.56236 -0.58068 O2 -0.63617 -0.69693 -0.59012 O3 -0.74727 -0.73899 -0.73981 N4 -0.65298 -0.65717 -0.60787 O5 -0.63987 -0.62519 -0.65063 Electrostatic Potential Map PCU-lactam PCU-lactim PCU-lactone The frontiers orbitals of PCU-models (a) HOMO for Lactam (b) LUMO for Lactam (a) HOMO for Lactim (b) LUMO for Lactim (a) HOMO for Lactone (b) LUMO for Lactone Polarizability (Å3), dipole moment (Debye) and Gibbs free solvation energy ∆Gsolv (kcal/mol) values of the PCU-models (Å3) µ (Debye) ∆GSolv PCU-models α PCU-lactam 81.218 6.4726 -17.925 PCU-lactim 81.663 3.5616 -17.085 PCU-lactone 79.324 1.2617 -8.893 (kcal/mol) Acknowledgements We thank the National Research Foundation for financial support, UKZN, and the CHPC (www.chpc.ac.za) for computational resources. Thank you for your kind attention