Recombination, Bacteriophages, and Horizontal Gene Transfer
advertisement

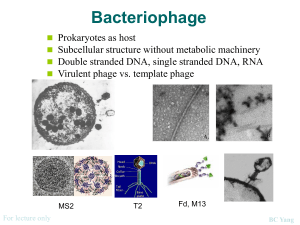
Horizontal Gene Transfer and Genetic Engineering Conjugation, Transformation, and Transduction Genetics Gene Transfer • Refers to the movement of genetic • • • information between organisms When genes are transferred between two bateria or a bacteria and a virus it involves a combination of the DNA from two different sources. This is referred to as recombination This type of transfer is referred to as horizontal( lateral) gene transfer Horizontal Gene Transfer • Horizontal gene transfer is a driving force in the development of drug resistance in bacteria • This type of transfer is different from the transmission of genetic chracteristics from one generation to generation vertically Gene transfer can occur between bacteria and plants • Agrobacterium tumefaciens lives in the soil • It is able to transfer a plasmid from its cells into • roots or stems through a scratch or injury to the plant tissue The plasmid integrates in host DNA and affects the host to cause the growth of tumors called CROWN GALLS Gene transfer from Bacteria to plant Gene transfer can occur between viruses and animals • • • • • SV 40 Simian virus Is a DNA virus Transforms or alters DNA and causes cancer Used to study cancer and HIV BCTERIAL CONJUGATION Bacterial Conjugation • transfer of DNA • • by direct cell to cell contact Contact with the pili discovered 1946 by Lederberg and Tatum + F x – F Mating • F+ = donor( contains the plasmid with the gene for conjugation) – This is referred to as the F factor or Fertility Factor • F– = recipient – does not contain F factor • F factor replicated by rolling-circle • • mechanism and duplicate is transferred ACROSS the pilus from the + to the recipients usually become F+ after it receives a copy of the DNA donor remains F+ Gene transfer and recombination • Genes are transferred in a linear manner • The F factor integrates into chromosomes at different points Mating • When two strains were mixed • There were incubated. • At intervals of 5 minutes, samples were taken of the F- cells • The cells were centrifuged so that they would know which genes were transferred. • The distance between genes was measured by the time that it took for the genes to be transferred. • During the first five minutes, the strains were mixed there was no recombination + F x – F • In its mating extrachromosomal state the factor has a molecular weight of approximately 62 kb Conjugative Proteins • Key players are the proteins that initiate the physical transfer of ssDNA, the conjugative initiator proteins • They nick the DNA and open it to begin the transfer • Working in conjunction with the helicases they facilitate the transfer of ss RNA to the F- cell - Formation of Hfr Hfr - high frequency of recombination DNA Transformation • Uptake of naked DNA molecule from the environment and incorporation into recipient in a heritable form • Competent cell – capable of taking up DNA • May be important route of genetic exchange in nature Transformation • Uptake of DNA can • • only occur at a certain cell density Cells need to be in the log phase of growth A competence factor is required for the uptake of DNA from the environment Streptococcus pneumoniae DNA binding protein competence-specific protein nuclease – nicks and degrades one strand Bacteria and transformation • Not all bacteria can be transformed in nature • Streptococcus pneumonia, Haemophilus influenza, and Neisseria gonorrhea Lab protocol Genetic recombination and transformation in the laboratory • Plasmids are designed to contain genes of interest • Transformation done in laboratory with species that are not normally competent (E. coli) • Variety of techniques used to make cells temporarily competent – calcium chloride treatment • makes cells more permeable to DNA Cloning vectors pAmp pGlo and transformation Microbial Genetics Bacteriophages Horizontal gene transfer • Clearly this plays a central role in the • • • • diversity of E. coli The greatest contributors are the bacteriophages Among the 18 prophage remnants on O157 – 12 resemble lambda phage They all contain a variety of deletions and or insertions Some of the phages are so similar that they contain a 20 kb segment tat is identical. Recombinant phages • It is believed that the phages have undergone • • • recombination and diversification They have been a major force in developing resistance and pathogenicity in bacteria such as E. coli and Streptococcus pyogenes Recombination could occur with in a single cell It could occur as the result of recombination Bacteriophages Bacteriophages • • • • Bacterial viruses Obligate intracellular parasites Inject themselves into a host bacterial cell Take over the host machinery and utilize it for protein synthesis and replication T- 4 Bacteriophage • Ds DNA virus • 168, 800 base pairs • Phage life cycles studied by Luria and Delbruck Bacteriophage structure Bacteriophage structure(con) • Most bacteriophages have tails • The size of the tail varies. • It is a tube through which the nucleic acid is injected as a result of attachment of the bacteriophage to the host bacterium • In the more complex phages the tail is surrounded by a contractile sheath for injection of the nucleic acids Bacteriophage structure • Many bacteriophages have a base plate and tail fibers • Some have icosahedral capsids • M13 has a helical capsid Capsid • The base plate requires 12 protein products • The head or capsid requires 10 genes • The capside requires scaffolding proteins for assembly • DNA packaging a mysterious process • Many phages lyse their host cells at the end of the intracellular phase T even phages Luria and Delbruck • Four distinct periods in the release of phages from • • • • host cells Latent period- follows the addition of phage( no release of virions) Eclipse period – virions were detectable before infection and are now hidden or eclipsed Rise or burst period – Host cells rapidly burst and release viruses The total number of phages released can be determined by the burst size – the number of viruses produced per infected cell General Steps Steps in the life cycle • Adsorption of the virus to the host • This is mediated by tail fibers or some analagous structure • When the tail fibers make contact, the base plate settles to the surface • This connection which is maintianed by electrostatic attraction and the ions Mg++ and Ca++ Attachment • There is host specificity in the attachment and adsorption of the bacteriophage • There are receptors for the attachment. They vary from bacteria to bacteria • The receptors are on the bacteria for other purposes: the bacteriophages evolved to utilize them for their invasion T even phages - Injection • The phage sheath shortens from 24 rings to 12 rings • The sheath becomes shorter and wider • This causes the central tube to push through the bacterial cell wall Gp5 • The baseplate contains the protein gp5 with lysozyme activity which made aid in the penetration of the host Early Genes • E. coli RNA polymerase starts transcribing genes( phage genes) within minutes of entering the bacterial cell • The early m RNA direct the synthesis of proteins and enzymes that are needed for hostile tack over • Some early virus specific enzymes degrade host DNA to nucleotides so that virus DNA synthesis can commence Late mRNA • Phage structural structural proteins • Proteins that help with phage assembly • Proteins involved in cell lysis and release Release • When the bacteriophages are released from the bacteria they can lyse the bacterial cell and break it open • They can be released through the cell membrane Irreversible attachment • The attachment of the tail fibers to the bacterium is a weak attachment • The attachment of the bacteriophage is also accompanied by a stronger interaction usually by the base plate Sheath contraction • The irreversible binding results in the sheath contraction Injection • When the irreversible attachment has been made and the sheath contracts, the nucleic acid passes through the tail and enters the cytoplasm Phage Multiplication Cycle – Lytic phages • Lytic phages or virulent phages enter the bacterial cell, complete protein synthesis, nucleic acid replication, and then cause lysis of the bacterial cell when the assembly of the particles has been completed. Eclipse Period • The bacteriophages may be seen inside or outside of • • • the bacterial cells The phages take over the cell’s machinery and phage specific mRNA’s are made Early mRNA’s are generally needed for DNA replication Later mRNA’s are required for the synthesis of phage proteins Intracellular accumulation phase • The bacteriophage sub units accumulate in the cytoplasm of the bacterial cell and are assembled Lysis or Release Phase • A lysis protein is released • The bacterial cell breaks open • The viruses escape to invade other bacterial cells Plaque assay • Phage infection and lysis can easily be • • • detected in bacterial cultures grown on agar plates Typically bacterial cells are cultured in high concentrations on the surface of an agar plate This produces a “ bacterial lawn” Phage infection and lysis can be seen as a clear area on the plate. As phage are released they invade neighboring cells and produce a clear area Plaque assay Lambda and Plaques • The plaque produced by Lambda had a different appearance on the Petri Dish. • It is considered to be turbid rather than clear • The turbidiy is the result of the growth of phage immune lysogens in the plaque • The agar surface contains a ratio of about a phage /107 bacteria MOI • Average number of phages /bacterium • After several lytic cycles the MOI gets higher due to the release of phage particles Transduction • Transfer of bacterial genes by viruses • Virulent bacteriophages – reproduce using lytic life cycle • Temperate bacteriophages – reproduce using lysogenic life cycle Generalized transduction • http://www.cat.cc.md.us/courses/bio141 /lecguide/unit4/genetics/recombination /transduction/gentran.html • http://www.cat.cc.md.us/courses/bio141 /lecguide/unit1/control/genrec/u4fg21a .html Generalized transduction • E. coli phage P21 or P22. • As a part of the lytic cycle, the phage cuts the • • • • bacterial DNA into fragments This fragmentation prevents the expression of bacterial genes Nucleotides can be used to make phage DNA Occasionally these DNA fragments are about the same size as phage DNA They become mistakenly packaged into phage capsids in place of phage DNA Types of Lysogenic Cycle • The most common type is the classic model of the • • • • Lambda phage The DNA molecule is injected into a bacterium In a short period of time, after a brief period of transcription, an integration factor and a repressor are synthesized A phage DNA molecule typically a replica of the injected molecules is inserted into the DNA As the bacterium continue to grow and multiply and the phage genes replicate as part of the bacterial chromosome Temperate • A bacteriophage that can exist as a lytic or lysogenic phage is referred to as a temperate phage • A bacterium containing a full set of phage genes is a lysogen • The process of infecting a bacterial culture with a temperate phage is called lysogenization Immunization • A bacterial cell or lysogen cannot be reinfected by a phage of the same type • This is resistance to superinfection is called immunity • More than 90% of the bacteriophages are temperate • These are unable to produce bursts such as T4 and T7 Lysogenic Phage Lambda Phage • Temperate phage • Alternate life cycle • Ds DNA – linear then circularizes when it enters the host • 48,502 base pairs • Molecular biology workhorse – because of its life cycle Genes Lambda genes • 46 genes have been identified • 14 are non esswential to the lytic cycle • Only 7 are nonessential to both the lytic and lysogenic cycles Life cycle of λ Phage Latency • Lysogenic conversion can lead to virulence • Botulism, cholera,and diptheria toxins are encoded by prophages that convert their host into a pathogenic bacterium Terminology • LEGEND • att: an E.coli seqence for the "attachment" or integration of • • • • • • lambda's circular chromosome. oriC: E.coli's origin of Chromosome replication (given here for orientation only) gal: E.coli's gene for galactose utilization pe:prophage ends (site of integration) cos: joined sticky ends of vegetative DNA; sometimes called ve ("vegetative ends") int: gene for the enzyme integrase c: gene for lambda repressor to maintain lysogeny • Q: another gene concerned with lysogeny • h: the last of the many capsomer genes. Bacteriophages Specialized transduction Attachment site • The E. coli chromosome contains one site at which lambda integrates. The site, located between the gal and bio operons, is called the attachment site and is designated attB since it is the attachment site on the bacterial chromosome. • The site is only 30 bp in size and contains a conserved central 15 bp region where the recombination reaction will take place. • The structure of the recombination site was determined originally by genetic analyses and is usually represented as BOB', where B and B' represent the bacterial DNA on either side of the conserved central element Recombination site • The bacteriophage recombination site - attP • is more complex. It contains the identical central 15 bp region as attB. The overall structure can be represented as POP'. However, the flanking sequences on either side of attP are very important since they contain the binding sites for a number of other proteins which are required for the recombination reaction. The P arm is 150 bp in length and the P' arm is 90 bp in length. Integration • Integration of bacteriophage lambda requires one phage-encoded protein - Int, which is the integrase and one bacterial protein - IHF, which is Integration Host Factor. • Both of these proteins bind to sites on the P and P' arms of attP to form a complex in which the central conserved 15 bp elements of attP and attB are properly aligned. • The integrase enzyme carries out all of the steps of the recombination reaction, which includes a short 7 bp branch migration. Normal Excision Generalized Transduction • Any part of bacterial genome can be transferred • Occurs during lytic cycle • During viral assembly, fragments of host DNA mistakenly packaged into phage head – generalized transducing particle Generalized transduction Specialized Transduction • Also called restricted transduction • carried out only by temperate phages that have established lysogeny • only specific portion of bacterial genome is transferred • occurs when prophage is incorrectly excised Specialized transduction Figure 13.20 Figure 13.20 Recombination and Genome Mapping in Viruses • viral genomes can also undergo recombination events • viral genomes can be mapped by determining recombination frequencies • physical maps of viral genomes can also be constructed using other techniques Specialized transduction mapping • provides distance of genes from viral genome integration sites • viral genome integration sites must first be mapped by conjugation mapping techniques Recombination mapping • recombination frequency determined when cells infected simultaneously with two different viruses Figure 13.24