h 2 - Barley World
advertisement

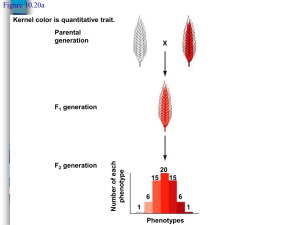
Qualitative and Quantitative traits • Phenotypes with discrete and easy to measure values. • Individuals can be correctly classified according to phenotype. • Show mendelian inheritance (monogene) • Little environmental effect • Molecular markers are qualitative traits • Examples: • Quantitative traits: • Individuals cannot be classified by discrete values • Quantitative trait distribution show a continuous range of variation and phenotypes can take any value • Complex mode of inheritance (polygene) • Moderate to great environmental effect) • Examples: Plant height, yield, disease severity, grain weight, etc % of plants • Qualitative traits: 20 30 Plant Height (in) 40 Inheritance of Quantitative traits The study of quantitative trait inheritance followed the same steps as for Mendelian traits. At the beginning they were thought to not follow Mendel’s laws. But it is not true F1 % of plants × PARENT 2: • pure line, completely homozygote • 20 inches F1: range of height distribution but no type of segregation P2 P1 PARENT 1: • pure line, completely homozygote • 40 inches 20 30 40 Plant Height (in) Plant Height (in) F2 % of plants F2: wider range of height distribution but no type of segregation 20 30 Plant Height (in) 40 Inheritance of Quantitative traits In 1903 the Danish botanist Wilhelm Johannsen measured the weight of seeds in the Princess variety of bean. This variety is a pure line since beans are self-fertilizing . Then he selected 19 beans of different weights and self-pollinated them several generations Doing this he got 19 pure lines (completely homozygous) in case they were not at the beginning of the experiment % of plants From a seed lot he measured and classified the beans by weight and obtained the range of distribution for that variety. 250 400 550 Weight (gr) He found that: The weight of the 5,494 beans he obtained followed a normal distribution All lines within each of the 19 groups were genetically identical but showed also a range of variation in weights. The average and distribution of weight in each pure line were similar to those of the original population Inheritance of Quantitative traits % of plants The Experiment of Johannsen 250 400 550 250 400 Weight (gr) 550 % of plants % of plants % of plants Weight (gr) 250 400 Weight (gr) 550 250 400 Weight (gr) Conclusions: •There is a genetic control that keeps the same average weight and distribution •However not all genetically identical seeds have the same weight. •The phenotype of each individual must be determined by the genotype and the environmental conditions •Without genetic variability, genetic improvement is not possible 550 Inheritance of Quantitative traits Johannsen showed that quantitative traits are determined by genes. However he did not find any type of mendelian segregation. This was studied in 1909 by Swedish Herman Nilsson-Ehle who studied kernel color in wheat He had several pure lines of red and white colored kernels. When crossing red x white he got always red F1, but different proportions of red and white kernels depending on the cross: a) 3 red : 1 white b) 15 red : 1 White c) 63 red : 1 white He deduced that the color was controlled by three loci. Only individuals with recessive homozygous alleles at the three loci showed the white phenotype. When a single dominant allele (A, B or C) is present at any of the three loci the red phenotype shows up. Inheritance of Quantitative traits a) 3 red : 1 white b) 15 red : 1 White c) 63 red : 1 white For case a), allelic variation between the two parents was present only at one locus P1 (red) P2 (white) AAbbcc X aabbcc F1(red) Aabbcc ¼ AAbbcc : ½ Aabbcc : ¼ aabbcc F2 (only one locus (red) (red) (white) segregating) Segregation 3 red : 1 white Inheritance of Quantitative traits a) 3 red : 1 white b) 15 red : 1 White c) 63 red : 1 white P1 (red) P2 (white) AABBcc X aabbcc F1(red) F2 (two loci segregating) AaBbcc 1/16 AABBcc 2/16 AABbcc 1/16 AAbbcc 2/16 AaBBcc 4/16 AaBbcc 2/16 Aabbcc 1/16 aaBBcc 2/16 aaBbcc 1/16 aabbcc (red) (red) (red) (red) (red) (red) (red) (red) (white) Segregation 15 red : 1 white Inheritance of Quantitative traits a) 3 red : 1 white b) 15 red : 1 White c) 63 red : 1 white P1 (red) P2 (white) AABBCC X aabbcc F2 (three loci segregating) 1/64 AABBCC 2/64 AABbCC 1/64 AabbCC 2/64AaBBCC 4/64 AaBbCC 2/64 AabbCC 1/64 aaBBCC 2/64 aaBbCC 1/64 aabbCC F1(red) AaBbCc Segregation 63 red : 1 white (red) (red) (red) (red) (red) (red) (red) (red) (red) 2/64 AABBCc 4/64 AABbCc 2/64 AabbCc 4/64 AaBBCc 8/64 AaBbCc 4/64 AabbCc 2/64 aaBBCc 4/64 aaBbCc 2/64 aabbCc (red) (red) (red) (red) (red) (red) (red) (red) (red) 1/64 AABBcc 2/64 AABbcc 1/64 Aabbcc 2/64 AaBBcc 4/64 AaBbcc 2/64 Aabbcc 1/64 aaBBcc 2/64 aaBbcc 1/64 aabbcc (red) (red) (red) (red) (red) (red) (red) (red) (white) Inheritance of Quantitative traits However, Nilsson-Ehle not only classified the seeds by color. He also classified them by color intensity and saw that color intensity also had a defined segregation pattern P1 (purple, X very dark red) P2 (white) F1(red) 1/16 : purple 4/16: dark-red 6/16: red 4/16: light-red 1/16: white He proposed that for this cross, color intensity was determined by two loci with two alleles each: one that produced red pigment (A and B) and other with no pigment (a and b). He determined that the effects of the alleles were additive and contributed equally to the phenotype, which depended on the number of alleles for pigment present P1 (purple, very dark red) P2 (white) AABB X aabb F1(red) AaBb 1/16 AABB 2/16 AABb 1/16 AAbb 2/16 AaBB 4/16 AaBb 2/16 Aabb 1/16 aaBB 2/16 aaBb 1/16 aabb (Purple) (dark-red) (red) (dark-red) (red) (light-red) (red) (light-red) (white) Inheritance of Quantitative traits P1 (purple, X very dark red) P2 (white) Frequency F1(red) Going one step further, He saw that within each of the groups there was also some variation - white + purple Color intensity 1/16 : purple 4/16: dark-red 6/16: red 4/16: light-red 1/16: white Inheritance of Quantitative traits He deduced that many loci were involved (not only two) in the trait and taking into account Johanssen’s findings: Phenotype=Genotype+Environment Frequency Then, the distribution of a quantitative trait would follow a normal distribution 4 + purple 3 1 2 - white Color intensity Analysis of quantitative traits is therefore complicated: Same genotype: 1 and 2 show different phenotype Same phenotype: 1, 3 and 4 is the result of three different genotypes Inheritance of Quantitative traits Frequency The inheritance of quantitative traits also explains the phenomenon of transgressive segregation: In the progeny of a cross we can get phenotypes out of the range of the parents P2 P1 0 Cold tolerance 10 Let’s assume 5 loci with additive effects control the trait P1 P2 aabbccddEE X AABBCCDDee F1 F2 AaBbCcDdEe All possible combinations of alleles at 5 loci. Between them: AABBCCDDEE (all favorable alleles) aabbccddee (all unfavorable alleles) Inheritance of Quantitative traits Quantitative traits are usually controlled by several genes with small additive effects and influenced by the environment Heritability h2 measures the proportion of phenotypic variation (variance) that is due to genetic causes P = G + E; h 2 VP = VG + VE VG VP A heritability of 40% for cold tolerance means that within that population, genetic differences among individuals are responsible of 40% of the variation. Therefore, 60% is due to environmental causes. However, that does not mean that the cold tolerance of a certain individual is due 40% to genetic causes and 60% to environmental causes. h2 is a property of the population and not of individuals Inheritance of Quantitative traits Heritability h2 measures the proportion of phenotypic variation (variance) that is due to genetic causes P = G + E; h 2 VP = VG + VE VG VP h2 ranges between 0 and 1 If h2 is 0 means : a) b) The trait is not genetically controlled. All the variation we see is due to environmental factors, or The trait is genetically controlled but all individuals have the same genotype h2 is very useful because it allows us to predict the response to artificial selection Inheritance of Quantitative traits Heritability h2 measures the proportion of phenotypic variation (variance) that is due to genetic causes VG 2 h P = G + E; VP = VG + VE VP h2 is very useful because it allows us to predict the response to artificial selection Frequency In plant breeding, the starting point is a segregating population (with genetic variability). The best individuals are selected to be the progenitors of the next generation μ0 Selection differential (S) = μS – μ0 μS Response to selection (R) = μR – μ0 0 Grain yield 6000 (lb/A) h Frequency μ0 μR 0 Grain yield (lb/A) Realized heritability: 6000 2 R S Is the ratio of the single-generation progress of selection to the selection differential of the parents. The higher h2, the higher the progress of selection in each generation Analysis of Quantitative traits The analysis of quantitative traits is based on the identification of the individual loci (QTL) controlling the trait, their location, effects and interactions A quantitative trait locus/loci (QTL) is the location of individual locus or multiple loci that affects a trait that is measured on a quantitative (linear) scale. These traits are typically affected by more than one gene, and also by the environment. Thus, mapping QTL is not as simple as mapping a single gene that affects a qualitative trait (such as flower color). Analysis of Quantitative traits There are two main approaches for QTL analysis: a) QTL analysis in mapping populations b) Association mapping Both approaches share a set of common elements: a) b) c) d) A population (array of individuals) that show variability for the trait of study Phenotypic information: We need to design an experiment to estimate the phenotypic value of each individual Genotypic information: A set of molecular markers that have been run in all the individuals of the population A statistical method to estimate QTL position, effects and interactions Analysis of Quantitative traits QTL analysis in mapping populations We need to develop a population from a single cross between two individuals that show contrasting phenotypes for the trait of study. For example, if we want to study quantitative resistance to Barley Stripe Rust (Puccinia striiformis f. sp. Hordei) we will develop a population from the cross between a susceptible line and a resistant line. The offspring of that cross will show recombination between the two parents and therefore, some individuals will be resistant and other will be susceptible Different types of mapping populations can be used: Doubled haploids (DH), Recombinant inbred lines (RIL), F2, Back cross (BC), etc. Always all individuals trace back to a single cross Analysis of Quantitative traits QTL analysis in mapping populations The first step is getting genotypic information for all the individuals of the population: molecular markers P2 Back Cross population P1 P2 SNP Parent 1 Parent 2 Line1 Line2 Line3 Line4 Line5 Line6 Line7 Line8 Line9 Line10 Line11 Line12 Line13 Line14 Line15 Line16 Line17 Line18 Line19 Line20 Line21 Line22 Line23 Line24 Line25 Line26 Line27 Line28 Line29 Line30 Line30 Line31 Line32 Line33 Line29 Line30 Line30 Line31 Line32 Line33 Line34 Line35 Line36 Line37 Line38 Line39 Line40 Line41 Line42 Line43 Line44 Line45 Line46 Line47 Line48 Line49 Line50 Line51 Line52 Line53 Line54 Line55 Line56 Line57 Line58 Line59 P1 1_0002 1_0004 1_0011 1_0014 1_0020 1_0023 1_0024 1_0026 1_0031 1_0036 1_0041 1_0047 1_0048 1_0050 1_0051 1_0052 1_0053 1_0055 1_0061 1_0063 1_0064 1_0065 1_0071 1_0073 1_0080 1_0081 1_0083 1_0084 G T A G C A T G G G G T T A T A A G T T T T G G T T G C A A T T G T A C C T T A A T A T T C G A C G C C G A C G G T T T C A A G G G G A T A A A A G T T T G C G T T G G A T A T C T A G C T T T A A T T T C G A C T G C T T C G G T T T C A T C G G T A T A A A A G T T T T C G T A G C G T T T C A A G G G G A T A A A A G T T T G C G T T G G G A A T G A T G C T T T A T T A A C T A T G G C G T C C G A T T C A T C C T T A A T A T T C G A T G C C G A C C G T A G G T T C G T G A T T T A A G G T C G G G T A G G A A T T C T A C G G T A T T A A A C G T C T C C G A C G G T T T G A A C G G T A T T T T T G T T T G G G T A G G G T A G G T A G G G T T T T T A A G T T C T G G T T G G A T T T C T T G C T T A A T A A A C G A C T C C T T C C A T T T C T T C C G G T A A T T T G G A C G G G T A G G G A T G C T T C G T T A T A A T T G T T C T C G G A G G A T T G C T T C C T T A A T T T T G T A C G G G T A G G G T T T C A A G G G G A T T T T T C T T T G G C T T C C A A A T G A A G C G T T A T T T T C T A T G G C G T C C G A A T G A T G C T T T A T T A A C T A T G G C G T C C G T T G C A T C C G G A A T A T T C T A T G C C T A C G A A A T C T A G G T T A T T A A A G G T C T C G G T G C G A A G C A T G C T G T A A A A A C T A T G C C G T C C G T A T G A T G C G T T A A T T T G T A T G G G T T G G G A A G G T A G C G G A A T T T T C G A C T G C G T C G A T A G G T T G G G T T T A A T T G G T C T C G T T G C A A A T G T A C G G T T A A A T T G G A C T C G G T G G A A A T G T A G G G T A A A A T T G G A C G C G G T G G G T T T C A A G G G G A T A A A A G T T T G C G T T G G G A T G C T T C C G T A T A A A A G T T C T C G G A G G A T T T C T A C G T T T T T A A A C T T C G C C T A C G A T T G G A T G G G T T T A A A A G G T T T C G T A G G A A T T C A A C G T T A T T T A A C G T T G G C G A C G G A A T G T T G C G T T A A A T T G T A C T C G G T G C A A A T C T A G C T T T A A A T T G G A C G C G G T G C A A T G G A T G C G T A A T A T T C T A T G C C G T C C G T A G C T A G G G G T A A T T T G G A C G G G T T G G A A A G G A A G G G G T T A T T T G T T T G G G G T G C A A T G C T A G C G G A A T A T T C G A C G C C G T C C G A A G G A A G G A A G G A A G G A A G G A A G G A A G A T T G C A T G A T T G C A T G A T T G C A T G A T T G A A T T C A T C A A T T C A T C A A T T C A T C A A T T G A A T C T A G G A A T C T A G G A A T C T A G G A A T G A A G G A A G G G G T T T T A A G G T T T G G G T C C A T T G C A T G C G T A A T T A A G T A T T G G T T G G A A T T C A T C C G T T A T A T T C T A T T C C G A C G G A A T C T A G G G G T T T A T T G T T C G C G G T G C A A A T G A T G G T G T T A T T T G T T T G G G G T G C G A T T G T A C C T G A A T A T T C T A C G C C G A C C G A T G G A A C G T T A T T A A A C T T T G C C G A C C G A T T G T T C C T T A A T A T T G G A C G C G G A G C High Throughput genotyping platform (SNP) G T A G G T T C C G T T A T T T T C G A T G G C T A C C A T T G C T T C C T T A A T T T T G T A C G G G T A G G A A A G G T A G C G T T T A T A A G G T C G G G G T G C A A A G G A A G C T T T A A A A A G T A T G C G G T G G G T A G C T A G G G G T A A T T T G G A C G G G T T G G G A A T G A T C C G T T A T T T T C T A T T G C G T C C A T T T C T A C C G T A A T T T T C T A C G G C T A C G G A A T C A A G C G G A A A T A A G T A T T G G G T G G A T A G C A T G G T T T T T A A A C T T T G C C T T C C G A T G C T T C G T T A T A T A A G G T C G G G G A G C G A A G C A A G G G T A T T A A A C T T T G C C G T C G A T A T G A A C C G G A A T T T T C T A T T G C T A C G A A T T C A A C G T T A T T T A A C G T T G G C G A C G A A A T G T A G C T T T T T T A A C G A C G G C G T C C G A A T C A T G C G T T A A T T T G G A T T G G G T G G G A T G C A A G C T G A A T T T T C T A T T G C G T C G G T T T C A A G G G G A T A A A A G T T T G C G T T G G A T T T C A T G C G G A A A T T T G G A T T G G T T G G Analysis of Quantitative traits QTL analysis in mapping populations If molecular markers are polymorphic, we can construct a linkage map based on recombination frequencies: 1H 0 7 12 18 22 25 26 29 30 36 48 54 58 61 68 73 86 87 96 101 111 119 121 122 130 133 136 2H BCD1434 DsT-66 Act8A RbgMD MWG837B scind00046 ABC165C Bmac0399 GBM1007 BCD098 GBM1042 BG367013 Bmag0211 BG369940 GBM1051 ABC160 JS10C Bmac0144A MWG706A KFP170 Blp ABC261 MWG2028 KFP257B WMC1E8 MWG912 ABG387A scssr04163 scssr08238 3H 0 5 7 17 DsT-1 ABG058 scind02622 ABG008 36 39 42 45 56 scssr10226 scssr07759 GBM1066 Pox scssr03381 scssr12344 scssr02236 Ebmac0684 BCD1434.2 ABG356 GBM1023 scsnp03343 vrs1 Bmag0125 DsT-41 MWG503 GBM1062 KFP203 MWG882A ABG1032 ABG072 Ebmc0415 cnx1 Zeo1 GBM1019 Aglu5F3R2 MWG720 GBM1012 wst7 scssr08447 MWG949A 63 65 68 71 83 88 94 97 102 103 104 108 117 124 137 139 149 161 163 165 170 173 179 180 0 4H BCD907 26 30 33 36 39 42 58 61 66 69 73 ABC171A GBM1074 scssr10559 MWG798B Dst-27 BCD706 DsT-39 alm Bmac0209 ABC325 DsT-67 87 89 98 scssr25691 ABG377 Bmag0225 121 124 125 Act8C ABG499 GBM1043 151 155 scsnp23255 ABG004 166 172 scind02281 MWG883 181 DsT-24 190 HVM62 199 DsT-40 212 218 ABC172 scssr25538 DsT-35 0 21 24 29 30 31 35 39 41 44 49 50 52 60 62 67 74 80 83 92 94 95 101 111 112 116 124 5H MWG634 MWG077 HVM40 DsT-29 CDO542 CDO122 hvknox3 Dhn6 ABC303 scssr20569 CDO795 HVM3 DST-46 scind03751 scssr18005 Tef2 GBM1020 Bmag0353 scind10455 DsT-79 scssr14079 ABG472 GBM1059 KFP221 Ebmac0701 MWG652B GBM1048 Hsh HVM67 KFP241.1 ABG601 6H 0 6 8 11 12 scssr02306 MWG618 DsT-6 ABC483 ABG610 37 44 45 53 55 56 58 ABG395 scssr02503 scssr18076 Bmac0096 NRG045A scsnp04260 Ale 79 82 85 90 100 ABC302 scind16991 scssr15334 scsnp06144 srh 111 scssr05939 120 128 134 141 RSB001A scsnp00177 0SU-STS1 ABG003B 157 166 169 170 179 193 197 198 205 207 215 223 224 225 scssr10148 Tef3 MWG877 BE456118A ABG496 scsnp02109 E10757A ABG391 JS10B ABC622 DsT-33 Bmag0113C MWG602A scssr03907 scssr03906 0 4 31 35 42 45 51 61 65 68 70 71 81 88 92 99 101 122 123 126 132 135 143 145 146 152 159 160 162 163 167 7H MWG620 Bmac0316 scssr09398 MWG652A MWG602B scind60002 JS10A GBM1021 GBM1068 BG299297 HVM31 rob Bmag0009 scssr02093 ABG474 Bmac0218C ABG388 scsnp21226 MWG820 GBM1008 scssr05599 MWG934 scind04312b scssr00103 GBM1022 Bmac0040 DsT-18 DsT-32B DsT-22 DsT-28 scind60001 DsT-74 MWG514 MWG798A DsT-71 0 14 20 29 36 38 44 57 66 68 69 73 82 86 97 98 103 115 117 125 126 127 137 139 ABG704 Bmag0007 scind00694 AW982580 MWG089 CDO475 ABG380 BE602073 scssr07970 scsnp00460 ABC255 ABC165D HvVRT2 scssr15864 GBM1030 scsnp22290 MWG808 DAK642 scind00149 scsnp00703 MWG2031 RSB001C nud lks2 ABC1024 Bmag0120 DsT-30 WG380B ABC310B Ris44 167 171 178 ABG461A WG380A GBM1065 196 197 199 HVM5 scssr04056 KFP255 ThA1 89 Analysis of Quantitative traits QTL analysis in mapping populations The basic QTL analysis method consists in walking trough the chromosomes performing statistical test at the positions of the markers in order to test whether there is a marker-trait association or not Disease severity (%) Parent 1(Resistant) Parent 2 (Susceptible) Line1 Line2 Line3 Line4 Line5 Line6 Line7 Line8 Line9 Line10 Line11 Line12 Line13 Line14 Line15 Line16 Line17 Line18 Line19 Line20 Line21 Line22 Line23 Line24 Line25 Line26 Line27 Line28 Line29 Line30 5 90 56 30 59 95 31 42 94 42 15 3 84 82 30 60 26 57 12 68 53 69 43 42 67 64 46 28 41 50 91 25 DsT-66 A B B A A A A A A A B B B B B A B B A A B B B A B B A A B B B B Analysis of 1H Quantitative traits Average Disease severy of plants with allele “A” (Inherited from Resistant parent) = 49.8 Average Disease severity of plants with allele “B” (Inherited from Susceptible parent) = 50.3 49.8 and 50.3 are not statistically different. Therefore, marker DsT-66 is not associated with resitance/susceptibility to the disease 0 7 12 18 22 25 26 29 30 36 48 54 58 61 68 73 86 87 96 101 111 119 121 122 130 133 136 BCD1434 DsT-66 Act8A RbgMD MWG837B scind00046 ABC165C Bmac0399 GBM1007 BCD098 GBM1042 BG367013 Bmag0211 BG369940 GBM1051 ABC160 JS10C Bmac0144A MWG706A KFP170 Blp ABC261 MWG2028 KFP257B WMC1E8 MWG912 ABG387A scssr04163 scssr08238 Disease severity (%) Parent 1(Resistant) Parent 2 (Susceptible) Line1 Line2 Line3 Line4 Line5 Line6 Line7 Line8 Line9 Line10 Line11 Line12 Line13 Line14 Line15 Line16 Line17 Line18 Line19 Line20 Line21 Line22 Line23 Line24 Line25 Line26 Line27 Line28 Line29 Line30 5 90 56 30 59 95 31 42 94 42 15 3 84 82 30 60 26 57 12 68 53 69 43 42 67 64 46 28 41 50 91 25 ABC261 A B B A B B A A B A A A B B A B A B A B B B A A B B A A A B B A Analysis of 1H Quantitative traits Average Disease severy of plants with allele “A” (Inherited from Resistant parent) = 30.4 Average Disease severity of plants with allele “B” (Inherited from Susceptible parent) = 69.8 30.4 and 69.8 are statistically different. Therefore, marker ABC261 is linked with a resitance/susceptibility QTL. The additive effect of the QTL is: a = (69.8-30.4)/2 = 14.7 0 7 12 18 22 25 26 29 30 36 48 54 58 61 68 73 86 87 96 101 111 119 121 122 130 133 136 BCD1434 DsT-66 Act8A RbgMD MWG837B scind00046 ABC165C Bmac0399 GBM1007 BCD098 GBM1042 BG367013 Bmag0211 BG369940 GBM1051 ABC160 JS10C Bmac0144A MWG706A KFP170 Blp ABC261 MWG2028 KFP257B WMC1E8 MWG912 ABG387A scssr04163 scssr08238 Significance trheshold MWG634 MWG077 HVM40 DsT-29 CDO542 CDO122 hvknox3 Dhn6 ABC303 scssr20569 CDO795 HVM3 DST-46 scind03751 scssr18005 Tef2 GBM1020 Bmag0353 scind10455 DsT-79 scssr14079 ABG472 GBM1059 KFP221 Ebmac0701 MWG652B GBM1048 Hsh HVM67 KFP241.1 ABG601 Probability 0 21 24 29 30 31 35 39 41 44 49 50 52 60 62 67 74 80 83 92 94 95 101 111 112 116 124 79 82 85 90 100 37 44 45 53 55 56 58 0 6 8 11 12 ABC302 scind16991 scssr15334 scsnp06144 srh ABG395 scssr02503 scssr18076 Bmac0096 NRG045A scsnp04260 Ale scssr02306 MWG618 DsT-6 ABC483 ABG610 RSB001A scsnp00177 0SU-STS1 ABG003B scssr05939 120 128 134 141 scssr10148 Tef3 MWG877 BE456118A ABG496 scsnp02109 E10757A ABG391 JS10B ABC622 DsT-33 Bmag0113C MWG602A scssr03907 scssr03906 111 157 166 169 170 179 193 197 198 205 207 215 223 224 225 0 4 31 35 42 45 51 61 65 68 70 71 81 88 92 99 101 122 123 126 132 135 143 145 146 152 159 160 162 163 167 Most likely position of the QTL CD907 BC171A BM1074 ssr10559 WG798B st-27 CD706 sT-39 m mac0209 BC325 sT-67 BC172 ssr25538 sT-35 sT-40 VM62 sT-24 ind02281 WG883 snp23255 BG004 ct8C BG499 BM1043 ssr25691 BG377 mag0225 Analysis of Quantitative traits QTL analysis in mapping populations Analysis of Quantitative traits 1H 0 7 12 18 22 25 26 29 30 36 48 54 58 61 68 73 86 87 96 101 111 119 121 122 130 133 136 2H BCD1434 DsT-66 Act8A RbgMD MWG837B scind00046 ABC165C Bmac0399 GBM1007 BCD098 GBM1042 BG367013 Bmag0211 BG369940 GBM1051 ABC160 JS10C Bmac0144A MWG706A KFP170 Blp ABC261 MWG2028 KFP257B WMC1E8 MWG912 ABG387A scssr04163 scssr08238 3H 0 5 7 17 DsT-1 ABG058 scind02622 ABG008 36 39 42 45 56 scssr10226 scssr07759 GBM1066 Pox scssr03381 scssr12344 scssr02236 Ebmac0684 BCD1434.2 ABG356 GBM1023 scsnp03343 vrs1 Bmag0125 DsT-41 MWG503 GBM1062 KFP203 MWG882A ABG1032 ABG072 Ebmc0415 cnx1 Zeo1 GBM1019 Aglu5F3R2 MWG720 GBM1012 wst7 scssr08447 MWG949A 63 65 68 71 83 88 94 97 102 103 104 108 117 124 137 139 149 161 163 165 170 173 179 180 0 4H BCD907 26 30 33 36 39 42 58 61 66 69 73 ABC171A GBM1074 scssr10559 MWG798B Dst-27 BCD706 DsT-39 alm Bmac0209 ABC325 DsT-67 87 89 98 scssr25691 ABG377 Bmag0225 121 124 125 Act8C ABG499 GBM1043 151 155 scsnp23255 ABG004 166 172 scind02281 MWG883 181 DsT-24 190 HVM62 199 DsT-40 212 218 ABC172 scssr25538 DsT-35 0 21 24 29 30 31 35 39 41 44 49 50 52 60 62 67 74 80 83 92 94 95 101 111 112 116 124 5H MWG634 MWG077 HVM40 DsT-29 CDO542 CDO122 hvknox3 Dhn6 ABC303 scssr20569 CDO795 HVM3 DST-46 scind03751 scssr18005 Tef2 GBM1020 Bmag0353 scind10455 DsT-79 scssr14079 ABG472 GBM1059 KFP221 Ebmac0701 MWG652B GBM1048 Hsh HVM67 KFP241.1 ABG601 6H 0 6 8 11 12 scssr02306 MWG618 DsT-6 ABC483 ABG610 37 44 45 53 55 56 58 ABG395 scssr02503 scssr18076 Bmac0096 NRG045A scsnp04260 Ale 79 82 85 90 100 ABC302 scind16991 scssr15334 scsnp06144 srh 111 scssr05939 120 128 134 141 RSB001A scsnp00177 0SU-STS1 ABG003B 157 166 169 170 179 193 197 198 205 207 215 223 224 225 scssr10148 Tef3 MWG877 BE456118A ABG496 scsnp02109 E10757A ABG391 JS10B ABC622 DsT-33 Bmag0113C MWG602A scssr03907 scssr03906 0 4 31 35 42 45 51 61 65 68 70 71 81 88 92 99 101 122 123 126 132 135 143 145 146 152 159 160 162 163 167 7H MWG620 Bmac0316 scssr09398 MWG652A MWG602B scind60002 JS10A GBM1021 GBM1068 BG299297 HVM31 rob Bmag0009 scssr02093 ABG474 Bmac0218C ABG388 scsnp21226 MWG820 GBM1008 scssr05599 MWG934 scind04312b scssr00103 GBM1022 Bmac0040 DsT-18 DsT-32B DsT-22 DsT-28 scind60001 DsT-74 MWG514 MWG798A DsT-71 0 14 20 29 36 38 44 57 66 68 69 73 82 86 97 98 103 115 117 125 126 127 137 139 ABG704 Bmag0007 scind00694 AW982580 MWG089 CDO475 ABG380 BE602073 scssr07970 scsnp00460 ABC255 ABC165D HvVRT2 scssr15864 GBM1030 scsnp22290 MWG808 DAK642 scind00149 scsnp00703 MWG2031 RSB001C nud lks2 ABC1024 Bmag0120 DsT-30 WG380B ABC310B Ris44 167 171 178 ABG461A WG380A GBM1065 196 197 199 HVM5 scssr04056 KFP255 ThA1 89 We identify the location of the QTL, the molecular markers flanking them, their effect and their interactions Analysis of Quantitative traits Association mapping Also called Linkage Disequilibrium mapping No need to develop populations from a single cross. Analysis is performed on arrays of related or unrelated individuals. Individuals of different origin, pedigree or degree of kinship may create population structure that can lead to false positives in the analysis. Association between markers and QTL in mapping populations are based only on linkage. However, in Association mapping these association can be due to multiple factors: linkage, selection, mutation, genetic drift, kinship, population structure, etc. Unlike mapping populations, where only alleles from the two parents are studied, multiple alleles may be present at any single locus. Analysis of Quantitative traits The analysis is based on the same principles as QTL analysis in mapping populations. Linkage maps are not needed SNP Line1 Line2 Line3 Line4 Line5 Line6 Line7 Line8 Line9 Line10 Line11 Line12 Line13 Line14 Line15 Line16 Line17 Line18 Line19 Line20 Line21 Line22 Line23 Line24 Line25 Line26 Line27 Line28 Line29 Line30 Line30 Line31 Line32 Line33 Line29 Line30 Line30 Line31 Line32 Line33 Line34 Line35 Line36 Line37 Line38 Line39 Line40 Line41 Line42 Line43 Line44 Line45 A higher density of markers is required 1_0002 1_0004 1_0011 1_0014 1_0020 1_0023 1_0024 1_0026 1_0031 1_0036 1_0041 1_0047 1_0048 1_0050 1_0051 1_0052 1_0053 1_0055 1_0061 1_0063 1_0064 1_0065 1_0071 1_0073 1_0080 1_0081 1_0083 1_0084 G T T T C A A G G G G A T A A A A G T T T G C G T T G G A T A T C T A G C T T T A A T T T C G A C T G C T T C G G T T T C A T C G G T A T A A A A G T T T T C G T A G C G T T T C A A G G G G A T A A A A G T T T G C G T T G G G A A T G A T G C T T T A T T A A C T A T G G C G T C C G A T T C A T C C T T A A T A T T C G A T G C C G A C C G T A G G T T C G T G A T T T A A G G T C G G G T A G G A A T T C T A C G G T A T T A A A C G T C T C C G A C G G T T T G A A C G G T A T T T T T G T T T G G G T A G G G T A G G T A G G G T T T T T A A G T T C T G G T T G G A T T T C T T G C T T A A T A A A C G A C T C C T T C C A T T T C T T C C G G T A A T T T G G A C G G G T A G G G A T G C T T C G T T A T A A T T G T T C T C G G A G G A T T G C T T C C T T A A T T T T G T A C G G G T A G G G T T T C A A G G G G A T T T T T C T T T G G C T T C C A A A T G A A G C G T T A T T T T C T A T G G C G T C C G A A T G A T G C T T T A T T A A C T A T G G C G T C C G T T G C A T C C G G A A T A T T C T A T G C C T A C G A A A T C T A G G T T A T T A A A G G T C T C G G T G C G A A G C A T G C T G T A A A A A C T A T G C C G T C C G T A T G A T G C G T T A A T T T G T A T G G G T T G G G A A G G T A G C G G A A T T T T C G A C T G C G T C G A T A G G T T G G G T T T A A T T G G T C T C G T T G C A A A T G T A C G G T T A A A T T G G A C T C G G T G G A A A T G T A G G G T A A A A T T G G A C G C G G T G G G T T T C A A G G G G A T A A A A G T T T G C G T T G G G A T G C T T C C G T A T A A A A G T T C T C G G A G G A T T T C T A C G T T T T T A A A C T T C G C C T A C G A T T G G A T G G G T T T A A A A G G T T T C G T A G G A A T T C A A C G T T A T T T A A C G T T G G C G A C G G A A T G T T G C G T T A A A T T G T A C T C G G T G C A A A T C T A G C T T T A A A T T G G A C G C G G T G C A A T G G A T G C G T A A T A T T C T A T G C C G T C C G T A G C T A G G G G T A A T T T G G A C G G G T T G G A A A G G A A G G G G T T A T T T G T T T G G G G T G C A A T G C T A G C G G A A T A T T C G A C G C C G T C C G A A G G A A G G A A G G A A G G A A G G A A G G A A G A T T G C A T G A T T G C A T G A T T G C A T G A T T G A A T T C A T C A A T T C A T C A A T T C A T C A A T T G A A T C T A G G A A T C T A G G A A T C T A G G A A T G A A G G A A G G G G T T T T A A G G T T T G G G T C C A T T G C A T G C G T A A T T A A G T A T T G G T T G G A A T T C A T C C G T T A T A T T C T A T T C C G A C G G A A T C T A G G G G T T T A T T G T T C G C G G T G C A A A T G A T G G T G T T A T T T G T T T G G G G T G C G A T T G T A C C T G A A T A T T C T A C G C C G A C C G A T G G A A C G T T A T T A A A C T T T G C C G A C C G A T T G T T C C T T A A T A T T G G A C G C G G A G C G T A G G T T C C G T T A T T T T C G A T G G C T A C C A T T G C T T C C T T A A T T T T G T A C G G G T A G G A A A G G T A G C G T T T A T A A G G T C G G G G T G C A A A G G A A G C T T T A A A A A G T A T G C G G T G G 4 1H-0-3_0969 1H-27.35-3_1276 1H-49.7-1_0159 1H-51.23-1_1484 1H-55.49-2_0798 1H-61.53-1_0798 1H-73.94-2_1126 1H-95.42-2_1373 1H-121.12-2_0908 1H-137.83-2_0138 2H-27.29-2_1015 2H-45.55-3_0363 2H-63.53-1_0191 2H-81.33-1_0859 2H-90.1-1_0969 2H-113.48-3_1402 2H-127.64-3_0310 2H-139.65-1_0551 3H-2.9-2_0159 3H-41-3_0953 3H-51.73-1_1313 3H-54.4-3_1008 3H-56.4-2_1062 3H-59.89-1_0373 3H-69.6-3_1242 3H-76.98-3_1346 3H-91.25-2_0659 3H-109.14-2_1513 3H-130.19-1_0280 3H-142.32-3_0137 3H-168.4-2_1267 4H-18.01-3_0150 4H-28.4-2_1374 4H-48.5-1_0577 4H-52.75-1_0946 4H-65.05-2_0906 4H-68.21-3_1536 4H-93.13-3_0142 4H-113.92-1_1066 5H-2.09-2_0226 5H-37.11-3_0410 5H-50.27-2_1308 5H-51-2_1011 5H-51.6-2_1260 5H-59.4-2_0961 5H-60.74-3_1280 5H-84.51-2_0096 5H-103.92-2_0327 5H-117.47-1_1200 5H-132.63-2_0259 5H-142.2-3_1366 5H-159.09-1_0820 5H-179.06-1_0254 6H-1.34-2_0881 6H-24.36-1_0868 6H-42.36-3_0783 6H-49.4-2_0291 6H-54.6-1_0962 6H-55.94-1_0513 6H-60.23-1_0270 6H-65.03-1_1261 6H-74.55-3_1088 6H-90.15-1_0202 6H-112.32-1_0239 6H-126.18-3_1498 7H-14.96-1_0841 7H-37.55-2_0126 7H-54.37-1_0772 7H-68.46-3_0639 7H-77.85-2_0879 7H-79.6-1_0370 7H-79.6-3_0835 7H-87.97-1_0143 7H-110.99-2_0385 7H-133.79-2_1104 7H-144.45-1_0843 Analysis of Quantitative traits 6 5 Significance threshold 3 2 1 0 Statistical test are performed at the position of each marker. The average phenotype of individuals with one genotypic class (with a certain allele) is tested against the average phenotype of individuals with other genotypic class (other allele) If differences between genotypic classes are statistically different, then there is marker-QTL association