11 - Brian Young, Ph.D.
advertisement
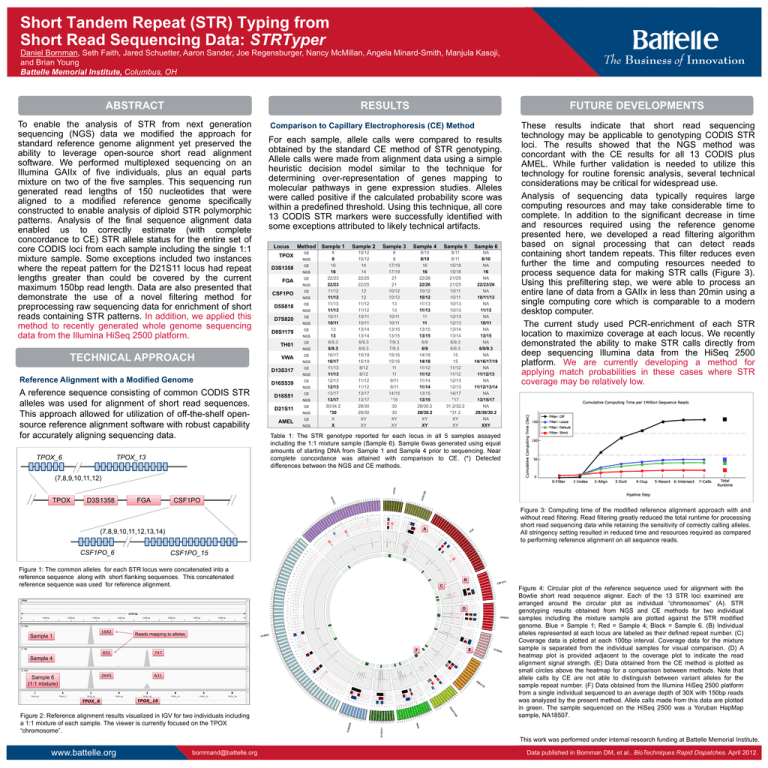
Short Tandem Repeat (STR) Typing from Short Read Sequencing Data: STRTyper Daniel Bornman, Seth Faith, Jared Schuetter, Aaron Sander, Joe Regensburger, Nancy McMillan, Angela Minard-Smith, Manjula Kasoji, and Brian Young Battelle Memorial Institute, Columbus, OH ABSTRACT RESULTS To enable the analysis of STR from next generation sequencing (NGS) data we modified the approach for standard reference genome alignment yet preserved the ability to leverage open-source short read alignment software. We performed multiplexed sequencing on an Illumina GAIIx of five individuals, plus an equal parts mixture on two of the five samples. This sequencing run generated read lengths of 150 nucleotides that were aligned to a modified reference genome specifically constructed to enable analysis of diploid STR polymorphic patterns. Analysis of the final sequence alignment data enabled us to correctly estimate (with complete concordance to CE) STR allele status for the entire set of core CODIS loci from each sample including the single 1:1 mixture sample. Some exceptions included two instances where the repeat pattern for the D21S11 locus had repeat lengths greater than could be covered by the current maximum 150bp read length. Data are also presented that demonstrate the use of a novel filtering method for preprocessing raw sequencing data for enrichment of short reads containing STR patterns. In addition, we applied this method to recently generated whole genome sequencing data from the Illumina HiSeq 2500 platform. Comparison to Capillary Electrophoresis (CE) Method For each sample, allele calls were compared to results obtained by the standard CE method of STR genotyping. Allele calls were made from alignment data using a simple heuristic decision model similar to the technique for determining over-representation of genes mapping to molecular pathways in gene expression studies. Alleles were called positive if the calculated probability score was within a predefined threshold. Using this technique, all core 13 CODIS STR markers were successfully identified with some exceptions attributed to likely technical artifacts. Locus TPOX D3S1358 Method Sample 1 Sample 2 Sample 3 Sample 4 Sample 5 Sample 6 CE 8 8 16 16 22/23 22/23 11/12 11/12 11/13 11/13 10/11 10/11 13 13 6/9.3 6/9.3 16/17 16/17 11/13 11/13 12/13 12/13 13/17 13/17 30/34.2 *30 X X 10/12 10/12 14 14 22/25 22/25 12 12 11/12 11/12 10/11 10/11 13/14 13/14 6/9.3 6/9.3 15/19 15/19 8/12 8/12 11/12 11/12 13/17 13/17 28/30 28/30 XY XY 8 8 17/19 17/19 21 21 10/12 10/12 13 13 10/11 10/11 13/15 13/15 7/9.3 7/9.3 15/16 15/16 11 11 9/11 9/11 14/15 *15 30 30 XY XY 8/10 8/10 16 16 22/26 22/26 10/12 10/12 11/13 11/13 11 11 13/15 13/15 6/9 6/9 14/18 14/18 11/12 11/12 11/14 11/14 13/15 13/15 28/30.2 28/30.2 XY XY 8/11 8/11 15/18 15/18 21/25 21/25 10/11 10/11 10/13 10/13 12/13 12/13 13/14 13/14 6/9.3 6/9.3 15 15 11/12 11/12 12/13 12/13 14/17 *17 31.2/32.2 *31.2 XY XY NA 8/10 NA 16 NA 22/23/26 NA 10/11/13 NA 11/13 NA 10/11 NA 13/15 NA 6/9/9.3 NA 14/16/17/18 NA 11/12/13 NA 11/12/13/14 NA 13/15/17 NA 28/30/30.2 NA XXY NGS CE NGS FGA CE NGS CSF1PO CE NGS D5S818 CE NGS D7S820 CE NGS D8S1179 CE NGS TH01 CE NGS TECHNICAL APPROACH VWA CE NGS D13S317 CE NGS Reference Alignment with a Modified Genome A reference sequence consisting of common CODIS STR alleles was used for alignment of short read sequences. This approach allowed for utilization of off-the-shelf opensource reference alignment software with robust capability for accurately aligning sequencing data. TPOX_13 TPOX_6 FUTURE DEVELOPMENTS D16S539 CE NGS D18S51 CE NGS D21S11 CE NGS AMEL CE NGS These results indicate that short read sequencing technology may be applicable to genotyping CODIS STR loci. The results showed that the NGS method was concordant with the CE results for all 13 CODIS plus AMEL. While further validation is needed to utilize this technology for routine forensic analysis, several technical considerations may be critical for widespread use. Analysis of sequencing data typically requires large computing resources and may take considerable time to complete. In addition to the significant decrease in time and resources required using the reference genome presented here, we developed a read filtering algorithm based on signal processing that can detect reads containing short tandem repeats. This filter reduces even further the time and computing resources needed to process sequence data for making STR calls (Figure 3). Using this prefiltering step, we were able to process an entire lane of data from a GAIIx in less than 20min using a single computing core which is comparable to a modern desktop computer. The current study used PCR-enrichment of each STR location to maximize coverage at each locus. We recently demonstrated the ability to make STR calls directly from deep sequencing Illumina data from the HiSeq 2500 platform. We are currently developing a method for applying match probabilities in these cases where STR coverage may be relatively low. Table 1: The STR genotype reported for each locus in all 5 samples assayed including the 1:1 mixture sample (Sample 6). Sample 6was generated using equal amounts of starting DNA from Sample 1 and Sample 4 prior to sequencing. Near complete concordance was attained with comparison to CE. (*) Detected differences between the NGS and CE methods. (7,8,9,10,11,12) TPOX D3S1358 FGA CSF1PO A (7,8,9,10,11,12,13,14) CSF1PO_6 Figure 3: Computing time of the modified reference alignment approach with and without read filtering. Read filtering greatly reduced the total runtime for processing short read sequencing data while retaining the sensitivity of correctly calling alleles. All stringency setting resulted in reduced time and resources required as compared to performing reference alignment on all sequence reads. CSF1PO_15 Figure 1: The common alleles for each STR locus were concatenated into a reference sequence along with short flanking sequences. This concatenated reference sequence was used for reference alignment. B C D 1682 Sample 1 Reads mapping to alleles 852 747 2645 631 F Sample 4 Sample 6 (1:1 mixture) TPOX_8 TPOX_10 Figure 2: Reference alignment results visualized in IGV for two individuals including a 1:1 mixture of each sample. The viewer is currently focused on the TPOX “chromosome”. E Figure 4: Circular plot of the reference sequence used for alignment with the Bowtie short read sequence aligner. Each of the 13 STR loci examined are arranged around the circular plot as individual “chromosomes” (A). STR genotyping results obtained from NGS and CE methods for two individual samples including the mixture sample are plotted against the STR modified genome. Blue = Sample 1; Red = Sample 4; Black = Sample 6. (B) Individual alleles represented at each locus are labeled as their defined repeat number. (C) Coverage data is plotted at each 100bp interval. Coverage data for the mixture sample is separated from the individual samples for visual comparison. (D) A heatmap plot is provided adjacent to the coverage plot to indicate the read alignment signal strength. (E) Data obtained from the CE method is plotted as small circles above the heatmap for a comparison between methods. Note that allele calls by CE are not able to distinguish between variant alleles for the sample repeat number. (F) Data obtained from the Illumina HiSeq 2500 platform from a single individual sequenced to an average depth of 30X with 150bp reads was analyzed by the present method. Allele calls made from this data are plotted in green. The sample sequenced on the HiSeq 2500 was a Yoruban HapMap sample, NA18507. This work was performed under internal research funding at Battelle Memorial Institute. www.battelle.org bornmand@battelle.org Data published in Bornman DM, et al.. BioTechniques Rapid Dispatches. April 2012.