Welcome to Part 2 of Bio 219
Lecturer – David Ray
Contact info:
Office hours – 1:00-2:00 pm MWTh
Office location – LSB 5102
Office phone – 293-5102 ext 31454
E-mail – david.ray@mail.wvu.edu
Lectures and other resources are available online at
http://www.as.wvu.edu/~dray.
Go to ‘Courses’ link
Chapter 10:
The Nature of the Gene and the
Genome
Inheritance
• Observation: Offspring resemble their
parents
– Question: How does this come about?
• Innumerable potential explanations can
be proposed:
– Homunculi?
– Components of sperm and egg mix like
paint?
– Are gametes and chromosomes involved?
The Gene
• A review of Gregor Mendel’s work
– Goal: to determine the pattern by which inheritable
characteristics were transmitted to the offspring
– Four major conclusions
Mendelian Inheritance
• Named for Gregor Mendel
– 1822-1884
– Studied discrete (+/-, white/black) traits in pea
plants
Mendelian Inheritance
• A classic experiment
• What did it tell Mendel?
– What conclusions can be drawn?
– Pod color was inherited as a discrete trait,
inheritance was not ‘blended’ for this trait
– Organism characteristics may be carried
as discrete ‘factors’ (now known as
‘genes’)
Mendelian Inheritance
• By continuing the experiment,
more can be observed
– The trait that was ‘lost’ in the first
generation (F1) was regained by the
second (F2), but in smaller numbers
• yellow + yellow = yellow and green
– The ‘factors’ come in different
versions (alleles)
– ‘Factors’ can mask one another –
dominant/recessive – but they are not
destroyed
– Further support for the discrete gene
hypothesis
Mendelian Inheritance
• By continuing the experiment,
more can be observed
– There was a definite mathematical
pattern to the occurrence of the traits
(3:1) in F2
– Comparison with mathematics
suggests that each offspring inherits
one allele from each parent (2 total)
– The phenotype (appearance) of the
plants was determined by the
genotype (actual combination of
alleles)
Mendelian ‘Model’ of Inheritance
• The true-breeders had two copies of one
type of allele (homozygous)
• Each parent passes on one of the alleles to
the offspring randomly
• The first generation will all be heterozygous
(have two different alleles)
• One of the alleles is able to block the other
(is dominant vs. being recessive)
• The F1’s pass on both of their alleles
randomly
• Simple math provides the expected ratios
of phenotypes and genotypes
The Gene
• A review of Gregor Mendel’s work
– Goal: to determine the pattern by which inheritable
characteristics were transmitted to the offspring
– Four major conclusions
–
–
–
1. Characteristics were governed by distinct units of inheritance (genes)
• Each organism has 2 copies of gene that controls development for each trait, one from
each parent
• The two genes may be identical to one another or nonidentical (may have alternate
forms or alleles)
• One of the two alleles can be dominant over the other and mask recessive alleles
when they are together in same organism
2. Gametes (reproductive cells) from each plant have only 1 copy of the gene for each trait;
plants arise from union of male & female gametes
3. Law of Segregation - an organism's alleles separate from one another during gamete
formation and are carried in that organism’s gametes.
Mendelian Inheritance
• Mendel’s results held true
for other plants (corn,
beans)
• They can also be
generalized to any
sexually reproducing
organism including
humans
Mendelian Inheritance
• Simple Mendelian inheritance
– Attached earlobes
– PTC (phenylthiocarbamide) tasting
– ‘uncombable hair’
• Complex (multigenic) inheritance
– Eye color
– Height
• Studying inheritance in humans is difficult
for ethical reasons but more easily done in
other organisms
Mendelian Inheritance
• Humans don’t typically have families large enough to see
mendelian ratios
• Inheritance can be tracked through the use of pedigrees
• Are the traits in white and black dominant or recessive?
Mendelian Inheritance
BB
bb
Bb
Bb
Bb
Bb
Bb
Bb
bb
Bb
bb
bb
bb
Bb
• If the trait indicated in
black is dominant we
would expect the cross
Bb
between 2 and 3 to
produce either ~50%
black trait and ~50%
white trait offspring or
100% black trait
offspring
Bb
• That ain’t the case
Mendelian Inheritance
bb
BB
Bb
Bb
Bb
Bb
Bb
Bb
• If the trait indicated in
black is recessive we
would expect the cross
between 2 and 3 to
produce all white trait
offspring
• Although it is possible
for individual 3 to have
a Bb genotype, it is
unlikely
• What is the genotype of
#2’s sister?
Mendelian Inheritance
• Using the information from the previous slides we can
deduce most individual’s genotypes
Bb
BB
B?
bb
Bb
B?
B?
Bb
Bb
bb
Bb
Bb
Bb
Bb
Bb
Bb
Bb
bb
Bb
bb
bb
bb
bb
Bb
bb
bb
Mendelian Inheritance
• The examples above are referred to as
monohybrid crosses since they deal with
only one trait at a time
• Mendel also followed dihybrid crosses in
which two traits are followed at once
• Would the traits segregate as a single unit
or independently?
Mendelian Inheritance
• A dihybrid cross
Mendelian Inheritance
• A dihybrid cross produced all
possible phenotypes and
genotypes
• Thus, all of the alleles behaved
independently of one another
• Mendel’s Law of Independent
Assortment – Each pair of alleles
segregates independently from
other pairs during gamete
formation
The Gene
• A review of Gregor Mendel’s work
– Goal: to determine the pattern by which inheritable
characteristics were transmitted to the offspring
– Four major conclusions
–
–
–
–
1. Characteristics were governed by distinct units of inheritance (genes)
• Each organism has 2 copies of gene that controls development for each trait, one from
each parent
• The two genes may be identical to one another or nonidentical (may have alternate
forms or alleles)
• One of the two alleles can be dominant over the other and mask recessive alleles
when they are together in same organism
2. Gametes (reproductive cells) from each plant have only 1 copy of the gene for each trait;
plants arise from union of male & female gametes
3. Law of Segregation - an organism's alleles separate from one another during gamete
formation and are carried in that organism’s gametes.
4. Law of Independent Assortment - segregation of allelic pair for one trait has no effect on
segregation of alleles for another trait. (i.e. a particular gamete can get paternal gene for
one trait & maternal gene for another)
Clicker Question
• Like most elves, everyone in Galadriel’s family has pointed ears (P),
which is the dominant trait for ear shape in Lothlorien. Her family
brags that they are a “purebred” line. She married an elf with round
ears (p), which is a recessive trait. Of their 50 children (elves live a
long time), three have round ears.
• What are the genotypes of Galadriel and her husband?
• ♀ = Galadriel; ♂ = husband
• A. ♀ PP; ♂PP
• B. ♀ pp; ♂ pp
• C. ♀ PP; ♂ Pp
• D. ♀ Pp; ♂ pp
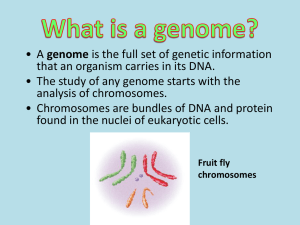
Chromosomes
• Mendel made no effort to describe what carried the genes, how
they were transmitted, or where they resided in an organism
•
1880s – Chromosomes are discovered because :
–
–
–
•
1. Improvements in microscopy led to…
2. observing newly discernible cell structures..
3. and the realization that all the genetic information needed to build & maintain a complex
plant or animal had to fit within the boundaries of a single cell
Walther Flemming observed:
–
–
1. During cell division, nuclear material became organized into visible threads called
chromosomes (colored bodies)
2. Chromosomes appeared as doubled structures, split to single structures & doubled at next
division
– Were chromosomes important for inheritance?
Chromosomes
• Are chromosomes important for inheritance?
– Hypothesis: If chromosomes are important for reproduction and
inheritance, altering the number of chromosomes delivered to offspring
should screw up the process.
– Theodore Boveri (German biologist) - studied sea urchin eggs fertilized
by two sperm (polyspermy) instead of the normal one single sperm
• 1. Disruptive cell divisions & early death of embryo
• 2. Second sperm donates extra chromosome set, causing abnormal cell
divisions
• 3. Daughter cells receive variable numbers of chromosomes
• Conclusion - normal development (reproduction/inheritance) depends upon a
particular combination of chromosomes & that each chromosome possesses
different qualities
Chromosomes
• Are chromosomes important for inheritance?
– Do chromosomes carry the genes?
– Whatever the genetic material is, it must behave in a manner consistent
with Mendelian principles
– Hypothesis: If chromosomes carry the genes necessary for inheritance,
they should mimic the theoretical behavior of genes
• Two copies per organism, Discrete units, Segregate independently into gametes
Chromosomes
• Are chromosomes important for inheritance?
– Hypothesis: If chromosomes carry the genes necessary for inheritance,
they should mimic the theoretical behavior of genes
• Two copies per organism, Discrete units, Segregate independently into gametes
– Experimental observations:
– Egg & sperm nuclei had two chromosomes each before fusion; Somatic
cells had 4 chromosomes
– Walter Sutton (1903) – Studied grasshopper sperm formation and
observed:
– 23 chromosomes (11 homologous chromosome pairs & extra accessory
(sex chromosome))
– 2 different kinds of cell division in spermatogonia
• mitosis (spermatogonia make more spermatogonia)
• meiosis (spermatogonia make cells that differentiate into sperm)
Chromosomes
• Are chromosomes important for inheritance?
– Haploid vs. Diploid
•
•
•
•
Haploid – having a single complement of chromosomes in a cell
Diploid – having a double set of chromosomes in a cell
Humans gametes? Human somatic cells?
23 chromosomes, 46 chromosomes
Chromosomes
• Are chromosomes important for inheritance?
– Hypothesis: There must be some mechanism to divide up the
chromosomes in the formation of gametes
– Experimental observations:
– Meiotic division (only observed in the formation of gametes) includes a
reduction division during which chromosome number was reduced by
half
– Two different kinds of cell division in spermatogonia
• mitosis (spermatogonia make more spermatogonia)
• meiosis (spermatogonia make cells that differentiate into sperm)
• If no reduction division, union of two gametes would double chromosome
number in cells of progeny
• Double chromosome number with every succeeding generation
Chromosomes
• In meiosis, members of each pair
associate with one another then
separate during the first division
• This explained Mendel's proposals
that :
– hereditary factors exist in pairs that
remain together through organism's life
until they separate with the production of
gametes
– gametes only contain 1 allele of each
gene
– the number of gametes containing 1 allele
was equal to the number containing the
other allele
– 2 gametes that united at fertilization
would produce an individual with 2 alleles
for each trait (reconstitution of allelic
pairs)
– Law of segregation
A a
AA aa
AA aa
A A
Aa
a
a
Chromosomes
• What about Mendel’s Law of
Independent Assortment?
– Having traits all lined up on a
chromosome suggests that they
would assort together, not
independently….
– as a linkage group
– Experiments in Drosophila
showed that most genes on a
chromosome did assort
independently… how?
– Is there some mechanism to
allow neighboring genes to assort
independenty?
Human
chromosome 2
Chromosomes
• What about Mendel’s Law of
Independent Assortment?
– Hypothesis: If neighboring genes
on a chromosome can assort
independently, there must be
some observable mechanism to
separate them
Human
chromosome 2
Chromosomes
• What about Mendel’s Law of
Independent Assortment?
– Hypothesis: If neighboring genes
on a chromosome can assort
independently, there must be
some observable mechanism to
separate them
– Experimental observations:
– 1909 – homologous chromosomes
wrap around each other during
meiosis
– During this process there is
breakage & exchange of pieces of
chromosomes
– Crossing-over and
recombination
Chromosomes
Typically, several cross-over events will occur between
well-separated genes on the same chromosome. Therefore,
genes E and F or D and F are no more likely to be co-inherited
than genes on different chromosomes.
Genes that are very close together (A and B), on the other hand,
are less likely to have cross-over events occur between them.
Thus, they will often be co-inherited (linked) and do not
strictly follow the Law of Independent Assortment.
Chromosomes
• Hypothesis: If the frequency of
independent assortment is related
to physical distance on the
chromosome, we can predict how
close two genes are by measuring
frequency of recombination.
• Since the likelihood of alleles
being inherited together is
influenced by their proximity…
• Genetic maps were possible by
determining the frequency of
recombination between traits
Clicker Question
• Three genes (1, 2, and 3) are present on a chromosome. The
recombination frequencies between them are:
• 1-2 = 11%
• 1-3 = 2%
• 2-3 = 13%
• Which diagram best approximates the relative locations of the genes
on the chromosome?
A.
1
2
3
B.
C.
D. 1 2
2
1
2
3
1 3
3
Chemical Nature of the Gene
• What is the genetic material?
• Observations:
• Chromosomes are likely the carriers
• Chromosomes consist primarily of three components
• Protein, RNA and DNA
• Are any of these the genetic material?
Chemical Nature of the Gene
• Which one (DNA, RNA or protein) is the actual genetic
material?
• Let’s narrow it down by hypothesis and experimentation
– Early experiments had shown that pneumonia causing bacteria that are
normally nonvirulent (R; rough) can be ‘transformed’ into the virulent (S;
smooth) type by some ‘transforming factor’ – the likely genetic material
Rough
Smooth
Chemical Nature of the Gene
• What was the ‘transforming’ or ‘genetic material’?
• Hershey and Chase (1952) – ‘blender experiment’
• Observations:
• Phage viruses consist of only two chemical
components – DNA and protein
• When a virus infects a cell, the cell makes many new
virus particles
• Thus, genes must enter the cell and direct it to make
new virus particles
• Which one enters the cell and actually becomes a
part of the new viruses?
Chemical Nature of the Gene
• What was the ‘transforming’ or ‘genetic material’?
• Avery et al. 1944 set up a multi-level hypothesis
• Extracted and separated DNA, RNA, and protein from
smooth (S; virulent) bacteria
• Three hypotheses:
• If protein is the genetic material, combining S-derived protein
with R bacteria will transform the R bacterial into the S strain
• If DNA is the genetic material, combining S-derived DNA with R
bacteria will transform the R bacterial into the S strain
• If RNA is the genetic material, combining S-derived RNA with R
bacteria will transform the R bacterial into the S strain
• Experimental observation:
• Only DNA was able to transform the strains
Chemical Nature of the Gene
• Label the phosphates in
DNA radioactively (32P) –
no phosphate in the protein
• Label the sulfur in the
protein (35S) – no sulfur in
the DNA
• Hypothesis: If the DNA
enters the cell, we should
find 32P in the infected cells
but not 35S (and vice versa)
• Observation: 32P in the
infected cells
• Animation online
Chemical Nature of the Gene
• Review of nucleic acid
structure:
– Phosphate
– Sugar
• Ribose or deoxyribose
– Nitrogenous base
•
•
•
•
Purines
Adenine and Guanine
Pyrimidines
Cytosine andThymine/Uracil
Chemical Nature of the Gene
• Review of nucleic acid
structure:
– Observation: Chargaff’s rules
– [A] = [T], [G] = [C]
– [A] + [T] ≠ [G] + [C]
– Suggested base pairing to Watson
and Crick, who later went on to
describe the overall structure of
DNA in vivo
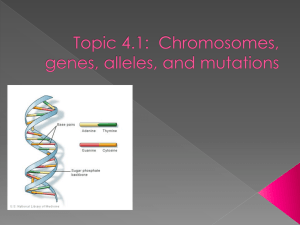
Chemical Nature of the Gene
• Review of nucleic acid
structure:
– Sugar-phosphate backbone
– Nitrogenous base rungs
– Directional – 5’ to 3’
Genome Structure
• Genome – the complete genetic complement
of an organism; the unique content of genetic
information
•
•
•
Early experiments to determine the structure of the genome
took advantage of the ability of DNA to be denatured
Denaturation – separation of the double helix by the addition
of heat or chemicals
How to monitor this separation?
• DNA absorbs light at ~260nm
• ss DNA absorbs more light, dsDNA less light
Clicker Question
• Which of the following 12 bp double helices will denature most
quickly?
A. 5’-AATCTAGGTAC-3’
3’-TTAGATCCATG-5’
C. 5’-AATTTAGATAT-3’
3’-TTAAATCTATA-5’
B. 5’-GGTCTAGGTAC-3’
3’-CCAGATCCATG-5’
D. They are all DNA, they will all
denature at the same rate.
Genome Structure
• DNA renaturation (reannealing) – the reassociation of
single strands into a stable double helix
• Seems unlikely give the size of some genomes but it
does happen.
• What does renaturation analysis allow?
•
•
•
Investigations into the complexity of the genome
Nucleic acid hybridization – mixing DNA from different organisms
Most modern biotechnology – PCR, northern blots, southern blots,
DNA sequencing, DNA cloning, mutagenesis, genetic engineering
Genome Structure
• Genome complexity - the variety & number of DNA
sequence copies in the genome
• Renaturation kinetics – what determines renaturation
rate?
• Ionic strength of the solution
• Temperature
• DNA concentration
• Incubation length
• Size of the molecules
Genome Structure
• Complexity in bacterial and viral genomes
A Cot curve uses the
Concentration and time
necessary for a genome
to renature to characterize
a genome
Simple genomes have
simple Cot curves
• MS-2 virus – 4000 bp genome
• T4 virus – 180,000 bp genome
• E. coli – 4,500,000 bp genome
Why do the smaller genomes
renature more quickly?
Genome Structure
• Complexity in eukaryotic genomes
• Eukaryotic Cot curves are more complex because the
genomes consist of different fractions
Genome Structure
• Complexity in eukaryotic genomes
• Highly repetitive DNA – Satellite DNAs - ~1-10% of
eukaryotic genomes
• Identical or nearly identical, tandemly arrayed
sequences
• Minisatellites – 10 – 100 bp repeats
• 5’- ATCAAATCTGGATCAAATCTGGATCAAATCTGG-3’
• Microsatellites – 1 – 10 bp repeats
• 5’-ATCATCATCATCATCATCATC-3’
Genome Structure
• Complexity in eukaryotic genomes
• Highly repetitive DNA – the
importance of satellite DNA
•
•
Centromeric DNA – the sections of
chromosomes essential for proper cell
division are mostly microsatellite DNA
DNA fingerprinting utilizes polymorphic microand minisatellite DNA – CODIS loci
Genome Structure
• Complexity in eukaryotic genomes
• Repeat expansion and human pathogenicity
•
•
•
•
•
•
CAG expansion in the huntingtin gene is associated with severity of
Huntington’s disease
CAG expansion produces long runs of glutamates in proteins
Polyglutamate chains tend to aggregate.
Inverse relationship between CAG repeat size and severity of
disease.
Normal range = (CAG)6 – (CAG)39
Disease range = (CAG)35 – (CAG)121
Genome Structure
• Complexity in eukaryotic genomes
• Moderately repetitive DNA – 10-80% of eukaryotic
genomes
• Coding repeats – Ribosomal RNA genes
• rRNA is necessary in large amounts
• Genes are arrayed tandemly
• Noncoding repeats – Interspersed aka mobile aka
transposable elements
• ~1/2 of your genome
• More on these later
Genome Stability
• Eukaryotic genomes are very dynamic over long
and short periods of time
• Whole genome duplication aka polyploidization
• offspring are produced that have twice the number of
chromosomes in each cell as their diploid parents
• May occur in either of two ways:
•
•
Two related species mate to form a hybrid organism that contains
the combined chromosomes from both parents (occurs most
often in plants)
Single-celled embryo undergoes chromosome duplication but
duplicates are not separated into separate cells, but are retained
in single cell that develops into viable embryo (most often in
animals)
Genome Stability
• Whole genome duplication aka polyploidization
• Polyploidization provides HUGE evolutionary
potential
• "extra" genetic information can:
- be lost by deletion
- be rendered inactive by deleterious mutations
- evolve into new genes that possess new functions
Genome Stability
• Gene duplication - duplication of a small portion of a
single chromosome
• Much more common than whole genome duplication
• Thought to occur most often via unequal crossover
•
•
Misalignment of chromosomes during meiosis
Genetic exchange causes one chromosome to acquire an extra
DNA segment (duplication) & the other to lose a DNA segment
(deletion)
Genome Stability
• Gene duplication – the globin cluster in primates
• Hemoglobin consists of 4 globin polypeptides
• (2 pairs: 1 pair always in ά-family, 1 in β-family)
• combinations differ with developmental stage
(embryonic, fetal, adult)
Transposable Elements and the Genome
– Transposable elements are sequences that
are interspersed throughout all eukaryotic
genomes examined.
– They play a role in the structure, function, and
evolution of the genome
Transposable Elements and the Genome
– Imagine a sequence that can copy itself and
then insert that copy somewhere else in the
genome
– What would expect to find when you line
some of them up?
Transposable Elements and the Human Genome
– Types of transposable elements
• Class I
– Retrotransposons
– LINEs, SINEs, SVA, LTR, ERV
– Defined as having an RNA intermediate
• Class II
– DNA transposons
– Mariner, hAT, piggyBac
– Defined as having a DNA intermediate
• Class II elements – cut and paste mobilization
• http://www.public.iastate.edu/~jzhang/Transposition.html
Generating Genetic Variation:
Normal SINE mobilization
Reverse transcription
and insertion
Pol III transcription
1. Usually a single ‘master’ copy
2. Pol III transcription to an RNA intermediate
3. Target primed reverse transcription (TPRT) – enzymatic machinery
provided by LINEs
Mobile Element Insertions and Mutation
Promoter
alters gene
expression
disrupts
reading
frame
disrupts
splicing
no disruption
ALU INSERTIONS AND DISEASE
LOCUS
BRCA2
Mlvi-2
DISTRIBUTION
de novo
de novo (somatic?)
SUBFAMILY
Y
Ya5
de novo
Familial
Ya5
Yb8
about 50%
Ya5
Familial
Y
Familial
one Japanese family
Ya5
Yb8
familial
Ya4
C1 inhibitor
ACE
de novo
about 50%
Y
Ya5
Factor IX
2 x FGFR2
GK
a grandparent
De novo
?
Ya5
Ya5
NF1
APC
PROGINS
Btk
IL2RG
Cholinesterase
CaR
Sx
DISEASE
Breast cancer
Associated with
leukemia
Neurofibromatosis
Hereditary desmoid
disease
Linked with ovarian
carcinoma
X-linked
agammaglobulinaemia
XSCID
Cholinesterase
deficiency
Hypocalciuric
hypercalcemia and
neonatal severe
hyperparathyroidism
Complement deficiency
Linked with protection
from heart disease
Hemophilia
Apert’s Syndrome
Glycerol kinase
deficiency
REFERENCE
Miki et al, 1996
Economou-Pachnis and
Tsichlis, 1985
Wallace et al, 1991
Halling et al, 1997
Rowe et al, 1995
Lester et al, 1997
Lester et al, 1997
Muratani et al, 1991
Janicic et al, 1995
Stoppa Lyonnet et al, 1990
Cambien et al, 1992
Vidaud et al, 1993
Oldridge et al, 1997
McCabe et al, (personal
comm.)
Genome Analysis
How many human genes?
80,000 Antequera F & Bird A, “Number of CpG islands and genes in
human and mouse”, PNAS 90, 11995-11999 (1993).
120,000 Liang F et al., “Gene Index analysis of the human genome
estimates approximately 120,000 genes”, Nat. Gen., 25, 239-240 (2000)
35,000 Ewing B & Green P, “Analysis of expressed sequence tags
indicates 35,000 human genes”, Nat. Gen. 25, 232-234 (2000)
28,000-34,000 Roest Crollius, H. et al., “Estimate of human gene number
Provided by genome-wide analysis using Tetraodon nigroviridis DNA
Sequence”, Nat. Gen. 25, 235-238 (2000).
41,000-45,000 Das M et al., “Assessment of the Total Number of Human
Transcription Units”, Genomics 77, 71-78 (2001)
Genome Analysis
• Sequencing a eukaryotic genome has become relatively
easy
• Figuring out what it all means is the hard part
• Human genome - ~25-30,000 genes (latest estimate)
• Nematode worm - ~25,000 genes
• Mustard plant - ~25,000 genes
• Puffer fish - ~25,000 genes
• What explains the differences in complexity and function
among different genomes?
• Comparative genomics suggests:
• Alternative splicing (more later)
• Differential regulation (more later)
Genome Analysis
• What explains the differences in
complexity and function among different
genomes?
• The protein-coding portion of the human
genome represents a remarkably small
percentage of total DNA (~1.1-1.6%)
•
•
The great majority of the genome consists
of DNA that resides between the genes &
thus represents intergenic DNA
Each of the 25-30,000 or more proteincoding genes consists largely of
noncoding portions (intronic DNA)
• How do we figure out what is a gene and
what isn’t?
Genome Analysis
• How do we determine what is important in a genome?
• Comparative genomics
• Conserved vs. nonconserved
• What are the “important” parts of a genome?
• Is most of the intergenic/intronic DNA subject to natural
selection?
• Are the intergenic/intronic portions conserved or nonconserved
• Protein coding and genetic control sequences tend to be ____.
…
…
Genome Analysis
• Comparative genomics
• The chimpanzee genome sequence was completed in 2005
• Much of what makes us human is likely to be determined through
finding differences between our genome and that of the chimp
Genome Analysis
• Comparative genomics
• FOXP2 a regulatory gene common to many vertebrates
• 2 amino acid differences are human specific (found only in
humans, not chimps or any other studied organism)
Genome Analysis
• Comparative genomics
• FOXP2 a regulatory gene common to many vertebrates
• Persons with mutations in FOXP2 gene suffer from a severe
speech & language disorder
• They are unable to perform the fine muscular movements of lips
& tongue that are required to engage in vocal communication
• Changes in FOXP2 that distinguish it from the chimp version
were fixed in human genome in the past 120,000 - 200,000
years; around the time modern humans may have emerged
Review of human genome complexity at:
http://www.dnalc.org/ddnalc/resources/chr11a.html