NAS Summer Institute 2010
Sassy Group 6: Gene Expression I
Harvard University
Tamara Brenner, Briana Burton, Robert Lue
University of California, Davis
Samantha Harris, Mitchell Singer
University of Iowa
Brenda Leicht, Bryant McAllister
University of
MinnesotaRobin Wright
(Facilitator)
University of Colorado, Boulder
Katie Chapman (Consultant)
NAS Summer Institute 2010
Sassy Group 6: Gene Expression I
Bryant
Katie
Mitch
Rob
Brenda
Briana
Samantha
Tamara
Sassy Group 6
Gene Expression I
NAS Summer Institute
June 25, 2010
Brenda, Briana, Bryant, Katie, Mitch,
Rob, Samantha, TamaraAnd Robin
Differential Regulation of Gene Expression
Determines Cell Form and Function
Level
Introductory Genetics
Introductory Biology (late in the course)
Knowledge of students prior to this unit
Know gene structure
Know the basic steps of the central dogma and
relevant techniques
Goals
Students will understand that:
Objectives
Students will be able to:
The information that allows regulation is
encoded in the DNA.
Define the regulatory components of a
gene.
Identify possible regulatory elements,
given a DNA sequence and/or protein
sequence.
Evaluate the functional significance of a
consensus sequence.
Protein presence, absence, varying levels
or activity can alter cell form or function.
Understand that this is important for
normal states (regulation) and can also
lead to abnormal states (misregulation).
Suggest how changes in gene expression
can lead to a disease state.
Make predictions about phenotypes
based on certain mutations.Organize
information on transcriptional components
into a regulatory circuit.
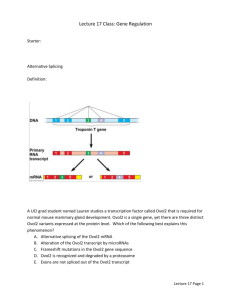
The Teachable Tidbit
Provides a hands-on activity for students to identify and examine
transcription factor binding sequences, and evaluate the effect of
variations in these sequences.
Forming a Consensus with Tidbits
7
Forming a Consensus
Blasts from the Recent Past
RNA polymerase transcribes DNA into RNA
HHMI Biointeractive
Blasts from the Recent Past
DNA-binding proteins initiate transcription
Drew Berry/HHMI
Blasts from the Recent Past
Different base pairs present
different patterns of hydrogen
bond donors and acceptors
adapted from Alberts et al
www.youtube.com/user/tomcfad
Genetic Control of Differences Between Females & Males
SRY
SRY
Chromatin Immunoprecipitation on Microarrays
(ChIP-on-chip):
a method to identify the DNA-binding sites of a
specific transcription factor
en.wikipedia.org/wiki/File:ChIP-on-chip_wet-lab.png
Group Activity: Identify the binding site for SRY
You are playing the roles of spots on a microarray chip. You are all 21-mer
nucleotide sequences discovered in this SRY ChIP-on-chip experiment.
Compare your sequence with others at your table and identify a series of
nucleotides in common.
Come to a consensus on the nucleotide sequence recognized by SRY.
TAAACGAAGGTAAACAATAGA
GCGCGCAACAATCCCGGGTTT
TAAACGAAGGTAAACAATAGA
TTCTTAACTAACAAACGCGAT
CCGTTTCACAACTATACTACT
AACAATAAGGCGACGCCTCTC
GCGCGCAACAATCCCGGGTTT
TACTTGATGGCAAACAATATA
TCGTTACGTAACAAACTCCAT
GGGTTTGACAACTATGCAATT
AACAATGAGGCGTTTTATCAC
AAGTGCAACAATAAGCCCTCT
TATAGGAAGCTAAACAATAGA
CCCTTAGCTAACTAACGCTAT
CTTTATGGCAACTATACTACT
AACAATACGGTTACGCCTATC
GGGCGCAACAATCACGGGTAT
Clicking for Data:
Forming a Consensus
Use your clicker to indicate the SRY binding site in your sequence.
A:
B:
C:
D:
AACTAT
AACAAT
AACAAA
AACTAA
AACAAT most common
AACTAT (-1)
AACAAA (-1)
AACTAA (-2)
Consensus
Group Discussion:
Forming a Consensus
Consensus
Group Discussion
Groups 1 and 2
Propose reasons why some sequences in your experiment are
more common than others.
Groups 3 and 4
Is it significant that these sequences have identical nucleotides at
certain positions but not at others? Defend your answer.
Groups 5 and 7
Predict the sequences to which the transcription factor binds
most strongly. How did you arrive at this conclusion?
http://www.picturingtolearn.org
Express and assess your
understanding through pictures
Picturing to Learn In-Class Activity
On a piece of paper you are going to create a freehand drawing
to explain the following to a high school senior taking biology:
How the degree of match with a consensus sequence can affect
the binding affinity of a transcription factor and the subsequent
level of gene expression.
Before you begin to draw, please think about the following: if
you were evaluating your drawing, what are the 3 most
important scientific components/concepts/ideas that need to be
included in your visual representation in order to explain the
science to your target audience? The most important should be
at the top.
Use a blank page to create your drawing. It should contain all the
necessary information you indicated above. Your audience will
not see your list, so your drawing should include some labels
and brief captions.
23
Back to SRY
(Sex determining Region Y Gene)
SRY is a transcription factor that regulates genes needed for development of
male-specific structures (e.g., testes) and male-specific traits
-SRY
+SRY
-SRY
+SRY
bio.miami.edu
SRY protein binds DNA
at the sequence
5’-ATAACAAT-3’
and bends the DNA in
that region to change
the openness of
chromatin.
Mutations in SRY or its Target Genes
• SRY Mutations
– Cause of 10-15% of cases of 46(X,Y) gonadal dysgenesis
– Some of these are nonsense or frameshift mutations that lead to
nonfunctional protein
– Many are missense mutations that affect the DNA binding ability of
the SRY protein.
• Target Genes
– MOA-A; altered expression is implicated in several
neuropsychiatric disorders including depression, autism and ADHD,
which differ in incidence between males and females.
Homework
A group of Russian scientists, Savinkova et al (2008), identified changes
in the TATA box of human promoters that lead to distinct pathologies.
Some of the mutations and resulting phenotypes are listed below.
Savinkova, L.K., et al: Biochemistry (Moscow)
2009, 74(2): 117-129.
Homework
1. Make a drawing that explains why some changes in the TATA box lead
to a decrease in protein expression, while others lead to an increase in
protein expression.
2. Imagine that you identify individuals with the mutation in the Globin promoter that is listed in the table. However, these individuals
express normal levels of -globin protein and are not affected by thalassemia. It turns out that these individuals have additional
mutations in their DNA. What possible mutation(s) could explain why
these individuals do not express the disease?
Goals
Students will understand that:
Objectives
Students will be able to:
The information that allows regulation is
encoded in the DNA.
Define the regulatory components of a
gene.
Identify possible regulatory elements,
given a DNA sequence and/or protein
sequence.
Evaluate the functional significance of a
consensus sequence.
Protein presence, absence, varying levels
or activity can alter cell form or function.
Understand that this is important for
normal states (regulation) and can also
lead to abnormal states (misregulation).
Suggest how changes in gene expression
can lead to a disease state.
Make predictions about phenotypes
based on certain mutations.Organize
information on transcriptional components
into a regulatory circuit.
30