NMPDRtoBench
advertisement

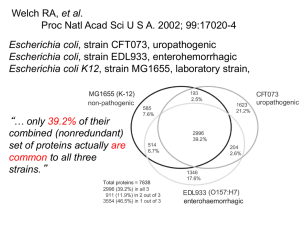
National Microbial Pathogen Data Resource Connecting Bioinformatics to the Bench Leslie Klis McNeil NCSA, University of Illinois, Urbana NMPDR is a BRC • NIAID Bioinformatic Resource Centers common goals different focus organisms • Provide annotations and tools to develop diagnostics and therepeutics against Priority Pathogens • NMPDR core organisms, all category B: Campylobacter jejuni Listeria monocytogenes Staphylococcus aureus Strepcococcus pyogenes and pneumoniae Vibrio cholerae, vulnificus, parahaemolyticus www.nmpdr.org Sister BRCs focus on other priority pathogens • Unified port of entry at • Eight BRCs curate viruses, protozoa, and bacteria, or insect vectors of disease www.nmpdr.org Who is NMPDR • Fellowship for Interpretation of Genomes Primary software developers Curators who do manual annotation • Computation Institute at University of Chicago Software developers Hardware managers • Argonne National Laboratory Software developers • NCSA University of Illinois at Urbana Education, outreach, training www.nmpdr.org What is NMPDR • Genome database with value added Manual annotation in context of systems biology Comparative analysis tools • • • • • Bidirectional Best Hits—select and align Functional clusters—genes with conserved proximity Compare regions—adjust size of region, number of genomes Pinned regions—phylogenetic comparison with all genomes Signature genes—find genes in common or that distinguish userselected groups of genomes; groups may contain one or many Essential genes page Drug target discovery and in silico screening Organism pages with phenotype information www.nmpdr.org Pathogen-specific gateways to data www.nmpdr.org Outreach services in the user interface • User forum links to iLabs with Inquiry Units for teaching and training • PathInfo—VBI’s PIML project, info about General info and strain descriptions Lab handling and safety Epidemiology • Journals button opens most recent, relevant ASM articles • Google news—RSS feed of popular press • Links to resources such as strain collections www.nmpdr.org Annotation Status Table • Immediate access to genes whose functions are known with some degree of certainty Named genes in subsystems Named genes not in subsystems Hypothetical genes in subsystems • Gateway to genes about which nothing is known Hypothetical genes not in subsystems • List of genes with links to NMPDR analysis tools • Exploration in comparative framework first step to formulating working hypotheses about functions www.nmpdr.org Pathways to Data • Start with keyword search for name of gene or protein • Start with sequence of your gene or protein and blast against any complete genome • Start by browsing an organism of interest View lists of proteins with/without functional names; included/not in biological subsystem. Choose one from the list to investigate with comparative tools. • Start from subsystems tree to view the phylogenetic distribution of an interesting biological process • Start from essential genes page to view essential genes in model organisms and to project essentiality to closely or distantly related organisms • Start from virtual structural proteomes to investigate proteins about which structural information is available in PDB www.nmpdr.org Subsystems approach to genome annotation • Subsystems annotation provides researchers with corrected functional annotations in a structured biological context • Consistency across genomes achieved by vertical annotation of functions rather than horizontal focus on single genomes • More than 500 distinct subsystems have been developed Metabolic pathways Complex structures Genotype – phenotype associations • Subsystems integrate genomic and functional contexts of genes in metabolic reconstructions or populated subsystem spreadsheets • Metabolic reconstructions summarize all subsystems in a given genome • Populated subsystems compare all genomes in a given subsystem www.nmpdr.org What is a Subsystem? • Subsystem is a generalization of pathway Collection of functional roles jointly involved in a biological process or complex • metabolic, signaling, regulatory, structural • Functional Role is the abstract biological function of a gene product Atomic or fundamental; examples: • 6-phosphofructokinase (EC 2.7.1.11) • LSU ribosomal protein L31p • cell division protein FtsZ www.nmpdr.org Expert-Defined Subsystems • Curator is researcher with first-hand knowledge of biological system • Functional roles defined and grouped into subsystem and subsets by curator universal groups of roles include all organisms functional variants are subsets of roles found in a limited number of organisms • often represent alternative paths www.nmpdr.org Populated Subsystems • Two-dimensional integration of functional roles with genomes universal groups of roles include all organisms functional variants are subsets of roles found in a limited number of organisms • Spreadsheet Columns of functional roles Rows of organisms Cells of annotated genes • Table of functional roles with GO terms • Diagram www.nmpdr.org Simple Example: Histidine Degradation Subsystem • Conversion of histidine to glutamate is organizing principle • Functional roles defined in table: Subsystem: Histidine Degradation 1 2 3 4 5 6 7 HutH HutU HutI Glu F HutG NfoD ForI Histidine ammonia-lyase (EC 4.3.1.3) Urocanate hydratase (EC 4.2.1.49) Imidazolonepropionase (EC 3.5.2.7) Glutamate formiminotransferase (EC 2.1.2.5) Formiminoglutamase (EC 3.5.3.8) N-formylglutamate deformylase (EC 3.5.1.68) Formiminoglutamic iminohydrolase (EC 3.5.3.13) www.nmpdr.org Subsystem Diagram • Three functional variants • Universal subset has three roles, followed by three alternative paths from IV to VI Tetrahy drofolate S u bsyste m Diagram Form im inotetrahy drofolate GluF NH 3 I HutH H2 O II H2 O HutU III Form am ide H2 O HutI IV H2 O VI NH 3 ForI www.nmpdr.org HutG V NfoD Subsystem Spreadsheet Subsystem Spreadsheet Organism Variant HutH HutU HutI GluF HutG NfoD Bacteroides thetaiotaomicron 1 Q8A4B3 Q8A4A9 Q8A4B1 Q8A4B0 Desulfotela psychrophila 1 gi51246205 gi51246204 gi51246203 gi51246202 Halobacterium sp. 2 Q9HQD5 Q9HQD8 Q9HQD6 Q9HQD7 Deinococcus radiodurans 2 Q9RZ06 Q9RZ02 Q9RZ05 Q9RZ04 Bacillus subtilis 2 P10944 P25503 P42084 P42068 Caulobacter crescentus 3 P58082 Q9A9MI P58079 Q9A9M0 Pseudomonas putida 3 Q88CZ7 Q88CZ6 Q88CZ9 Q88D00 Xanthomonas campestris 3 Q8PAA7 P58988 Q8PAA6 Q8PAA8 Listeria monocytogenes -1 • • • • • • Column headers taken from table of functional roles Rows are selected genomes, or organisms Cells are populated with specific, annotated genes Shared background color indicates proximity of genes Functional variants defined by the annotated roles Variant code -1 indicates subsystem is not functional www.nmpdr.org ForI Missing Genes Noticed by Subsystems Annotation • No genes were annotated “ForI (EC 3.5.3.13) Formiminoglutamic iminohydrolase” when the Histidine Degradation subsystem was populated • Organisms missing ForI convert His to Glu • Candidate genes that could perform the role “ForI” must be identified • Strategy for finding genes is based on chromosomal clustering and occurrence profiling www.nmpdr.org Finding Genes that Cluster with NfoD • Green gene is NfoD of Xanthomonas • Blue genes within 10 kb of NfoD in at least four other species • finds biggest clusters in other species • fc-sc shows table of homologous pairs in other genomes • displays homologous regions in other genomes www.nmpdr.org What are Pinned Regions? • Focus gene is number 1, colored red • Most frequently co-localized homolog numbered 2, colored green • Homologous genes presented in the same color with the same numerical label • Numerical labels correspond to rank ordered frequency of co-localization with the focus gene Focus gene labeled 1 Gene 17 is homolog 16th most frequently colocalized with focus gene www.nmpdr.org Candidate ForI in Context with NfoD • Homologous regions around NfoD, red, center • Same color indicates homology BLAST cutoff 1e-20 • HutH, the first functional role in the subsystem, is green, 2 • Candidate ForI is pink, 4, “conserved hypothetical” www.nmpdr.org Annotation of ForI EC 3.5.3.13 • Metabolic context proves need for role Organisms missing annotated ForI degrade His to Glu • Chromosomal context points to candidate Clusters with NfoD and other genes in subsystem • Occurrence context supports candidate Organisms containing NfoD lack GluF and HutG, required for functional variants 1 and 2, respectively Organisms containing candidate ForI also contain NfoD, indicating functional variant 3 • Phylogenetic trees of candidate ForI genes are coherent www.nmpdr.org Conjectures archived in HOPS • Hypotheses and Open Problems identified by Subsystems HOPS linked from NMPDR’s FAQ • Subsystems point to missing or alternative genes • Bioinformatic predictions need to be tested at the bench • ForI candidate now verified experimentally • Connections forged between bench and bioinformatics www.nmpdr.org Bioinformatics to Bench • Essential genes page at NMPDR Click bar to search for essential genes Follow NMPDR link to compare with other genomes www.nmpdr.org Candidate Drug Targets • First-draft table (manually derived) links to biochemical data in BRENDA or TCDB • Candidate proteins essential in at least one of the NMPDR pathogens included in subsystems by our curators orthologs in the Protein Data Bank orthologs in a substantial number of bacterial priority pathogens curated in the BRC system • Second-draft table to be automatically generated annotations include essential for growth or virulence PDB and pathogen orthologs No good hit in host targets without crystallized orthologs suggested to HTS project at Argonne National Laboratory www.nmpdr.org NMPDR efforts feed into high-throughput structure project at Argonne www.nmpdr.org In Silico Screening • Targets docked with 10 K random compounds as training set • Neural network program tracks 9 properties of compounds to learn characteristics of those that bind and those that do not • ZINC compound db screened to find 10K likely binders predicted to be ligands • Targets docked against 10K predicted ligands on BlueGene with Dock5 • Top 1000 docked compounds soon to be linked to NMPDR www.nmpdr.org IBM BlueGene Supercomputer World’s fastest Supercomputer 280 TeraFLOPS www.nmpdr.org Live Demo of NMPDR • From essential genes, click H.pylori, then click NMPDR for first protein • Show compare regions Possible to increase/decrease size of region Possible to “walk” chromosome Possible to include more genomes--type in 10 and click resubmit • Click on the homologous gene 1 in the second genome, Campylobacter • Ask, is this function also essential in Campy,is this a good drug target? • Investigate the campy homolog by using Pins, Compare Regions, find best clusters (CL) • What is the pathway or biological system that this protein is essential for? IF not included in a subsystem by NMPDR curators, follow alias link to KEGG • Pathway is lysine biosynthesis—Ask: Does this protein catalyze the rate-limiting step? Is this the best function in this pathway to target for inhibition by a drug? Does this protein have a close structural/functional homolog in human or PDB? Use BLAST to find homologs. Is this a broad or narrow spectrum target? Show all homologs using Bidirectional Best Hits button. www.nmpdr.org