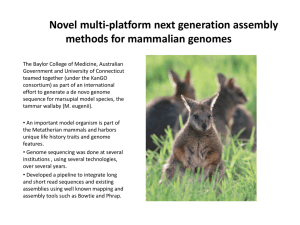
Assembly_reloaded
advertisement
COMPUTATIONAL GENOMICS GENOME ASSEMBLY Members: Eishita Tyagi Sandeep Namburi Aarthi Talla Vinay Vyas Amin Momin Jay Humphrey Contents • Assembly – De novo • Algorithms Involved – Reference – Assembly problems – Task and Strategy How do we get Reads? De novo Assembly Reads Overlap Local Multiple Alignment Alignment Scoring Contigs Scaffolding Finishing Assembly Problems: -Repeats -Chimerism -Gaps Overlapping Reads • Greedy Algorithm • Overlap-Layout-Consensus Algorithm • Eulerian path Algorithm Greedy Algorithm X = abcbdab Y = bdcaba, the lcs is Z= bcba. LCS = Longest common subsequence By inserting the non-lcs symbols while preserving the symbol order, we get the scs: = abdcabdab Shortest common superstring The union of two strings (X U Y) Overlap-Layout-Consensus Algorithm • Graph based: G(V,E) How is it executed ?? – – – – • • • de Bruijn Graph – a directed graph with vertices that represent sequences of symbols from an alphabet, and edges that indicate where the sequence may overlap. Nodes (V) = reads Edges (E) = between overlapping reads Path = Contig (each node occurs at least once) • Builds graph – alignments Removing ambiguities Output is a set of nonintersecting simple paths, each path being a contig. Consensus sequence • E.g.. Celera Assembler, Arachne Eulerian Path Algorithm • De-bruijn graph • Eulerian path – a path that visits all edges of a graph • Breaks reads into overlapping n-mers. • Source: n-1 prefix and destination is the n-1 suffix corresponding to an n-mer. – Build a table of n-mers contained in sequences (single pass through the genome) – Generate the pairs from n-mer table ATG AT TGC TG GCA GC n-mer CAG CA AGG AG GGT HAMILTONIAN (IDURY WATERMAN GG EULER MSA •Correct errors using multiple alignment •Score alignments •Accept alignments with good scores Parameters for Scoring • length of overlap • % identity in overlap region • maximum overhang size Contigs • A continuous sequence of DNA that has been assembled from overlapping cloned DNA fragments. • Reads combined into Contigs based on sequence similarity between reads. Scaffolding The process through which the read pairing information is used to order and orient the contigs along a chromosome is called Scaffolding. – Scaffolding groups contigs -> subsets with known order and orientation. – Nodes (V) = contigs. – Directed edge (E) – mate pairs between node. Mate Pairs or Paired End Reads • A library of Paired End reads or Mate pairs are used to determine the orientation and relative positions of contigs. • • • • • Reads sequenced from the template DNA Known order and orientation (facing in, facing out, or facing the same direction) between reads. Known range of separation between read 5' ends. Approximately 84-nucleotide DNA fragments that have a 44-mer adaptor sequence in the middle flanked by a 20-mer sequence on each side. Mate-pairs allow you to remove gaps & merge islands (contigs) into super-contigs. Sameward Outward Inward Mate Pairs are Needed to: •Order Contigs •Orient Contigs •Fill Gaps in the assembly A scaffold of 3 contigs (the thick arrows) held together by mate pairs Reference Assembly Reads Overlap Local Multiple Alignment Alignment Scoring Assembly Problems: -Repeats -Chimerism -Gaps Contigs Map to a reference Finishing Mapping contigs to a reference Assembly Problems • Errors from sequencing machines, e.g. missing a base, or misreading a base • Even at 8-10 X coverage, there is a probability that some portion of the genome remains unsequenced • Repeat problem lead to Misassembly and Gaps • Chimeric reads - When two fragments from two different parts of genome are combined together Repeat Problems • Ability of an assembly program to produce 1 contig for a chromosome: limited by regions of the genome that occur in multiple near-identical copies throughout the genome (repeats). • Assembler incorrectly collapses the two copies of the repeat leading to the creation of 2 contigs instead of 1. • Thus, number of contigs increase with the number of repeats. • Repeated sequences within a genome also produce problems with higher level ordering. Genome mis-assembled due to a repeat. Assembly programs incorrectly may combine the reads from the two copies of a repeat leading to the creation of 2 separate contigs (Contig Level Misassembly) Gaps • A good Assembler would have to ignore the repeats and generate one contig instead of two. • A Gap would be created in the place of the repeat. • Higher the number of repeats, the Gaps generated would increase. Chimeric reads •Two fragments from two different parts of genome are combined together. •Can give a completely wrong assembly. Finishing • Process of completing the chromosome sequence. • Re-sequence areas with gaps or less than 2x, 3x, 5x coverage • Close gaps (usually by PCR or BACs) • Expensive and time-consuming. Our Task • To Assemble Neisseria meningitidis strains sequences: M13519 and M16917 – Strains are Non-groupable • M13519 matches Serogroup C (PCR), W135 (SASG) • M16917 matches Serogroup Y (PCR), W135 (SASG) • No completed genomes available for strains with Serogroup Y and W135. Our Strategy De novo assembly with Newbler and Mira3 Reference assembly using AMOScmp and Newbler Best results from each merged with Best Minimus2 Finish by manual alignment Important Assembler Metrics • • • • • • • Number of large contigs Total size Coverage Average length N50 Longest contig % genome assembled NEXT PRESENTATION – WEDNESDAY Initial Results and Lab