Low-level (n=2)
advertisement

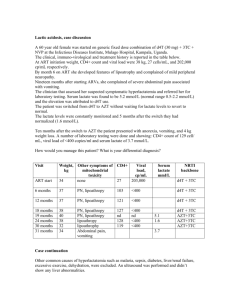
Emerging patterns of drug resistance and viral tropism in cART-naïve and failing patients infected with HIV-1 subtype C Thumbi Ndung’u, BVM, PhD Associate Professor Director, HIV Pathogenesis Programme Doris Duke Medical Research Institute Nelson R. Mandela School of Medicine University of KwaZulu-Natal 100 HIV-1 Phylogeny H 100 47.2% C 100 F1 100 100 F2 100 K 100 D 100 100 M-group B 100 100 100 100 100 100 100 12.3% J G A1 27.0% A2 N-group CPZ US CPZ GAB 100 O-group CPZ ANT 5% Phenotypic Classification of HIV-1 • Slow/low versus rapid/high • Syntitium-inducing (SI) versus NSI • Slow/Low = NSI (Early, slow progression) • Rapid/High = SI (Late, rapid progression) HIV-1 coreceptor usage and viral tropism Virus Variants M-tropic T-tropic Dual tropic CCR5 Macrophage CD4 CXCR4 Primary T cell CCR5 CD4 Target Cell Types CXCR4 T-cell line CD4 >25 years of HIV/AIDS > 33 For every 2 people put on treatment, 5 others are infected Selection of Resistant strains Treatment begins Drug-susceptible quasispecies Viral load Drug-resistant quasispecies Selection of resistant quasispecies Incomplete suppression •Inadequate potency •Inadequate drug levels •Inadequate adherence •Pre-existing resistance Time Study rationale Background: • Relatively limited information on coreceptor usage by HIV-1 subtype C isolates, particularly in children. However, most studies suggest very rare CXCR4 usage • Some reports suggest increasing X4 usage (in adults) eg. Johnston et. al. (n=28), 50% using X4 among ART experienced viremic patients • Previously used methods may be biased because they involved first generating viral isolates by co-culture Study rationale • ART may boost T-cell immune responses which have been shown to preferentially suppress X4 viruses. Thus partially effective therapy may select against X4 viruses (Deeks et al, JID 2004; Harouse et al, PNAS 2003) • ART reduces CCR5 expression on T cells (due to reduction in T cell activation) potentially selecting for X4 viruses (Brumme et al, JID 2005; Anderson et al, AIDS 1998) • Suboptimal drug metabolism (such as AZT) in the cellular reservoirs for X4 viruses has been suggested and could lead to selection for X4 viruses (Boucher et al, AIDS 1992) Aims Specific Aims: 1) To determine the prevalence of major drug mutations in ART-naïve and failing children and adults 2) Determine overall prevalence of X4 tropism among children and adults initiating and failing HAART 3) Compare prevalence of X4-utilizing viruses between ART-naïve and ART-experienced subjects with detectable viremia 4) Explore factors associated with viral tropism in HIV1C infection HIV-1 Genotyping Assay plasma centrifugation Blood cells RNA RT-PCR cDNA PCR DNA PCR Dye terminators A T T C G A T T C T G C C G Software analysis ATAGACCAG : consensus sequence I Q Q I Q *L ATCGACCTG : patient sequence Trofile assay summary- for coreceptor usage 5’LTR tat rev vif gag vpu pol Luc env 3’LTR vpr + pcDNA-env CMV pA Env Luciferase assay CCR5 cells 0.2µ filter 293T cells 0.2µ filter CXCR4 cells Table 1: Children Demographic and Clinical Characteristics Characteristics HAART-Failures (n=41) HAART-Naïve (n=40) P value 7.9 (4.8-10.4) 0.9 (0.5-2.8) <0.0001a Black Race 41 (100.0) 39 (97.5) 0.49b Male Gender 24 (58.5) 18 (45.0) 0.27b 9.0 (3.1-13.5) (n=33) 14.0 (7.5-22.0) (n=37) 0.008a Current CD4%, median (IQR) 18.0 (9.0-24.0) 14.0 (7.5-22.0) (n=37) 0.47a Current CD8%, median (IQR) 51.0 (40.5-58.0) 48.0 (35.5-56.5) (n=37) 0.38a Current CD3%, median (IQR) 72.0 (67.0-77.0) 66.0 (56.0-77.5) (n=37) 0.18a 4.9 (4.4-5.4) 5.9 (5.6-6.8) <0.0001a Age, median years (IQR) Nadir CD4%, median (IQR) Current plasma HIV-1 viral load, median log-10 copies/ml (IQR) Current WHO Stage: (n=40) I 1 (2.5) 0 (0.0) II 15 (37.5) 1 (2.5) 0.003b III 18 (45.0) 20 (50.0) IV 6 (15.0) 19 (47.5) NOTE. Data are no. (%) of children unless otherwise indicated. For cases where the data is incomplete, the n value is indicated. Statistical tests: a Mann-Whitney U test and b Fisher’s exact test (for WHO stage analysis, stages I, II and III were grouped together). Table 1: Patient Demographic and Clinical Characteristics Cont. Characteristics HAART-Failures (n=41) HAART-Naïve (n=40) P value Current Drug regimen: D4T, 3TC, EFV 25 (61.0) D4T, 3TC, LPV/r ● 6 (14.6) D4T, DDI, EFV * 1 (2.4) AZT, 3TC, NVP 3 (7.3) AZT, 3TC, EFV ○ 3 (7.3) AZT, DDI, EFV 1 (2.4) AZT, DDI, LPV/r 1 (2.4) D4T, ABC, LPV/r * 1 (2.4) Duration of HAART prior to study 28.6 (19.7-37.5) (n=38) recruitment, median months (IQR) History of single-dose NVP for PMTCT 10 (26.3) (n=38) 18 (47.4) (n=38) 0.09b NOTE. Data are no. (%) of children unless otherwise indicated. For cases where the data is incomplete, the n value is indicated. Prior treatment indicated with underlined drug/s changed ● d4T, 3TC, ritonavir (n=1); * unknown; ○ d4T, 3TC, EFV (n=1) and AZT, 3TC, NVP (n=1); d4T, 3TC, kaletra; d4T, 3TC, EFV. Statistical tests: a Mann-Whitney U test and b Fisher’s exact test Frequency of drug resistance mutations and levels of resistance in HAART-failing children to the NRTIs (a) and NNRTIs (b) 58.5% had TAMs 39% had ≥3 TAMs Average no. of major mutation in patients failing standard first line treatment (n=30) • d4T/3TC/EFV (n=25) – 3 patients have no DRMs (VLs are 617; 79,400; 228,000) – 20 NRTI DRM – 2 NNRTI DRM (one patient had a PI DRM) • d4T/3TC/kaletra (n=5) – 3 patients have no DRMs (VLs are 143,000; 198,000; 4,410,000) – 1 patient has 1 NRTI DRM (M184V) only – 1 patient has 1 NRTI (M184V) and 1 NNRTI DRM (Y181C) How many major mutations compromise the standard second line treatment? d4T/3TC/EFV (n=25) → AZT/ddI/Kaletra • 3 patients susceptible to all drugs – no change needed • All patients susceptible to kaletra • 3 patients susceptible to 3 drugs in standard second line tx. AZT Resistance ddI Resistance Susceptible (n=2) High-Level (n=2) Potential low-level (n=2) Low level (n=5) Low-level (n=1) Intermediate (n=2) Potential low-level (n=2) Intermediate (n=8) Low-level (n=3) Intermediate (n=2) High-level (n=1) High-Level (n=4) Intermediate (n=2) High-Level (n=2) • Overall, 13 of 25 (52%) patients will have some degree of resistance (low to high) to two of the three drugs in their new regimen (excluding potential low-level resistance) d4T/3TC/kaletra (n=5) → AZT/ddI/(NVP/EFV) • 4 of 5 patients are susceptible to all second line drugs • 1 patient had intermediate resistance to EFV (3.7 yrs old)(Y181C) Note: Overall better if not changed • All still susceptible to PIs and d4T with 3 patients still susceptible to 3TC [2 high-level resistance to 3TC (M184V)] Comparison of coreceptor usage in HAARTfailing and HAART-naïve children HAART-Naïve HAART-Failures HAART-Failures 9.4% 42.9% 42.9% 45.7% 45.7% R5-tropic D/M-tropic R5-tropic R5-tropic X4-tropic X4-tropic D/M-tropic D/M-tropic 90.6% 11.4% 11.4% p<0.0001 Evaluation of Several Genotypic Tools for the Prediction of CXCR4usage Prediction of CXCR4-usagea Genotypic Tool Sensitivity (%) Specificity (%) PPV (%) NPV (%) aA 11/25 charge rule 30.0 96.9 83.0 74.0 Net V3 charge rule 65.0 78.1 59.0 82.0 C-PSSMsinsi 75.0 87.5 75.0 88.0 Geno2pheno[coreceptor]b 60.0 87.5 70.0 82.0 Combined Rulesc 63.2 100.0 100.0 85.0 C4.5 25.0 100.0 100.0 73.0 C4.5 positions 8-12 25.0 100.0 100.0 73.0 PART 30.0 100.0 100.0 75.0 SVMwetcat 40.0 96.9 86.0 77.0 total of 52 pure subtype C isolates with both phenotypic and genotypic data were included in this analysis. bA false positive rate of 10% was used. c A combination of the first four genotypic tools were used where the majority prediction was considered as the final genotype prediction (n=47). Adult patient information Patient HAART-Experienced HAART-Naïve p-value Characteristic Patients failing Patients (n=45) Treatment (n=45) Age, median years (Q1-Q3) Gender: Female 36 (24-51) 36 (20-78) 28 (65%) 27 (60%) Black race 45 (100%) 45 (100%) 174 (9-718) 57 (3-197) 6, 653 (225-220,010) 123 (8-660) 44,042 (1,702-1,167,759) 32 (71 %) 13 (29 %) 9 (20 %) 36 (80%) CD4 count, median 3 cells/mm (Q1-Q3) Current Nadir Vial load, median copies/ml WHO stage at visit I-III IV 0.65 0.036 0.0004 0.001 Patterns of drug resistance • What is the outcome of patients failing if started on the standard second line of treatment without having genotypic data? Average no. of major mutation in patients failing standard first line treatment (n=16) • d4T/3TC/ (EFV/NVP) (n=16) (Note: 2 on NVP) – No major PI mutations – 1.75 NRTI DRM – 1.69 NNRTI DRM How many compromise the standard second line treatment? d4T/3TC/ (EFV/NVP) (n=16) → AZT/ddI/LPV/r • All patients susceptible to kaletra (LPV/r) • 6 patients susceptible to all 3 drugs in standard second line tx. AZT Resistance ddI Resistance Potential low-level (n=3) Susceptible (n=4) High-Level (n=1) Susceptible(n=1) Potential low-level (n=2) Low-level (n=1) Low-level (n=2) Low-level (n=2) Intermediate (n=2) Low-level (n=2) • 4 of 16 (25%) patients will have some degree of resistance (low to intermediate) to two of the three drugs in their new regimen (excluding potential low-level resistance). • 6 of 16 (37.5%) will have some degree of resistance (low to high) to one of the three drugs in their new regimen (excluding potential low-level resistance). High levels of CXCR4 viruses in patients failing therapy- limited salvage options V3 loop-based methods for coreceptor usage prediction Method 11/25 Overall net V3 charge C-PSSM Geno2Pheno Combined algorithm* % of sequences correctly predicted 78 75 81 84 87 % of R5 sequences correctly predicted 90 71 85 86 90 % of X4/D/M sequences correctly predicted 55 81 72 82 80 *In the combined algorithm, concordant results from at least 3 of 4 methods (i.e. the amino acids at positions 11 and/or 25, the overall net V3 charge, C-PSSM prediction and Geno2Pheno prediction) were used. Conclusions • Virologic failure is mainly due to DRMs • High levels of TAMs is source of concern- suggests subpotimal adherence and need for intensive monitoring • Higher levels of CXCR4 using viruses among HAART experienced patients- need to explore CCR5 antagonists as part of first-line/early treatment • Collectively, these data highlight the need for intensified adherence counselling and better HAART monitoring to maximize benefits. Acknowledgements UKZN Monogram Biosciences • • • • • • • Jacqueline Reeves • Yolanda Lie • Elizabeth Anton Taryn Green Ashika Singh Mohendran Archary Michelle Gordon Raziya Bobat Hoosen Coovadia McCord Hospital • Henry Sunpath • Richard Murphy Harvard University • Daniel Kuritzkes • Bruce Walker Funding • • • • IMPAACT Network, NIH Harvard University CFAR South African DST/NRF Hasso Plattner Foundation