If you haven`t done so already, REGISTER TO VOTE!
advertisement

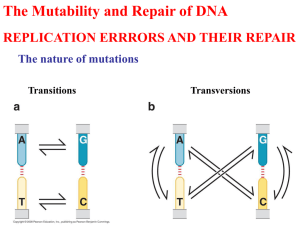
If you haven’t done so already, REGISTER TO VOTE! You live in a free country; make your voice heard! This website will take you through the process to register by mail in any of the 50 states: http://www.fec.gov/votregis/pdf/nvra.pdf States have different deadlines, so don’t delay Tests will be returned on Thursday, Sept. 23 in class Test answers will be posted on the website Thursday afternoon Gene Mutation and DNA Repair What is a mutation? A mutation is a change in the chemical composition of DNA Mutations Mutations are usually the source for genetic variation within populations. Mutations may be detrimental, beneficial or neutral. Mutations provide the basis for genetic studies. Types of Mutations 1. Chromosome abnormalities A. variation in number B. variation in arrangement 2. Alterations in base sequence at discrete locations Gametic vs. Somatic Mutations Somatic: expressed only in the mutated population; not passed on to subsequent generations Gametic: mutations that become part of the genetic makeup of the gametes and therefore can be passed on to subsequent generations Types of Mutations Dominant autosomal mutations: expressed in the first generation Recessive autosomal mutations: may not be expressed for many generations. Are usually only expressed in the absence of the dominant trait, and may be the result of crossing parents who each carry the recessive gene. Types of Mutations X-linked recessive: mutation associated with the Xchromosome. If the dam is homozygous (both chromatids have the same, recessive gene), the gene will be expressed in all male offspring (males inherit only one X-chromosome and receive it from the dam). Types of Mutations Lethal mutation: the organism cannot survive Conditional mutation: a mutation only expressed under certain environmental conditions e.g. temperature Causes of Mutations 1. 2. Spontaneous (random): arise independently of an identifiable external force. Induced: arise in response to a factor or force from or within the environment in which a cell or organism exists. Point Mutations Any change that disrupts a gene sequence or the coded information qualifies the change as a mutation. THE CAT SAW THE DOG Base Substitutions THE CAT SAW THE DOG THE CAT SAW THE HOG THE CAT SAQ THE DOG Frameshift: Deletion THE CAT SAW THE DOG THE ATS AWT HED OG C Frameshift: Insertion THE CAT SAW THE DOG M THE CMA TSA WTH EDO G How do mutations arise? Tautomers Structural isomers of nitrogenous bases (A,T, G and C) Shifts of a single proton Alter the bonding structure (base pairing) and therefore can induce changes in sequence during replication. Tautomers Note that the only change is a move of a hydrogen (proton) from one place on the ring to the next. The shifts are temporary, and the tautomers are much less stable than the normal forms, but while they exist, the tautomers can form abnormal base pairs Tautomers Base pairing between a tautomer and normal base will still be between a purine and pyrimidine, but will not preserve the information content. Thus, T will pair with G (three hydrogen bonds) A will pair with C (two hydrogen bonds) Tautomers Tautomers are transient because they are less stable, but if a base exists in its tautomeric form during replication, the wrong base can be incorporated into the replicated strand. Alkylating Agents Chemical agents that donate a –CH3 or CH2CH3 group to the keto groups (=O) of thymine and guanine Alter the base pairing properties Intercalating Agents Acridine orange, ethidium bromide Wedge between the bases and contort the DNA, increasing the probability of a slip during replication such that frameshift mutations are generated UV Radiation UV radiation causes the formation of thymine dimers Adjacent thymine residues on a strand of DNA become attached to one another 1. Unavailable to base pair with complement strand 2. Contorts the DNA strand 3. Inhibits normal replication Thymine Dimers High-Energy Radiation Shorter wavelength than UV Includes X-rays, gamma rays and cosmic radiation (ionizing radiation) Create free radicals that initiate chemical reactions that can damage DNA High-Energy Radiation 1. 2. Point mutations Break phosphodiester bonds of the DNA backbone (can induce chromosomal aberrations) For X-rays, the relationship between dose and mutation is linear High-Energy Radiation Cells in M (vs G1, S, or G2) are more susceptible because DNA is condensed into chromosomes and more accessible. (Cancer cells divide more frequently than normal cells, so this is why radiation can be an effective cancer treatment) Examples of conditions resulting from mutation or change in DNA sequence Duchenne Muscular Dystrophy arises most often from point mutations that alter the reading frame of the dystrophin gene. Examples of conditions resulting from mutation or change in DNA sequence ABO Blood groups Arise from differences in activity of the enzyme glycosyltransferase that modify substance H, a protein on the surface of red blood cells. A, B are different modifications of substance H O is the lack of modification of substance H Examples of conditions resulting from mutation or change in DNA sequence The O phenotype is due to a frameshift mutation (deletion) in the glycosyltransferase gene such that a non-functional protein is made The result is that substance H cannot be modified Ames test for mutagenicity Developed by Bruce Ames of the University of Iowa Asssess the ability of a compound or chemical to induce mutation Ames test for mutagenicity The test looks at the return mutation rate, that is, for the ability to grow in the absence of histidine (return to normal or wild type phenotype). Ames test for mutagenicity Uses strains of Salmonella bacteria that are sensitive to mutation (Protective coat and DNA repair mechanisms have been impaired) These strains are his -, that is, they cannot synthesize the amino acid histidine, and must be grown in the presence of histidine to survive. Ames test for mutagenicity The test compound is either incubated directly with bacteria, or Injected first into a mouse such that it passes through the liver to be modified in whatever way the compound would be metabolically processed if it entered the body (many compounds become more chemically reactive after passing through the liver) Ames test for mutagenicity The bacteria are then incubated on plates in medium that does not contain histidine. After 24 hours, the plates are removed and the number of bacterial colonies are counted Ames test for mutagenicity + histidine -histidine, Non-mutagenic compound -histidine, Mutagenic compound DNA Repair Numerous mechanisms exist to identify and repair mismatches and mutations Excision Repair: cut and paste 1. 2. 3. 4. Distortion or error is detected Nuclease clips out the error and several nucleotides on either side DNA polymerase fills in the gap with nucleotides, based on the nucleotide sequence of the complementary strand DNA ligase seals the nicks between the new DNA and the existing DNA strand at the 3’ end Excision Repair Xeroderma Pigmentosum (XP) People with XP (usually) lack the excision repair system and as such are very susceptible to skin damage Very susceptible to skin cancer Must be protected from UV radiation Proofreading and Mismatch Repair The normal error rate of DNA polymerase III (responsible for DNA replication in E coli) is 1 base per 100,000 bases replicated or 10-5. The E coli genome is 4 x 106 bases, so in the absence of proofreading, there would be 40 mutations every generation. Proofreading DNA pol III (and its eukaryotic compatriot) have proofreading functions, which means that the enzyme detects errors, reverses direction, clips out the wrong nucleotide and inserts the correct one. This reduces the error rate to 10-7, or one in ten million. Mismatch Repair 10-7 is still pretty high, so a second mechanism exists to further reduce errors in replicated DNA. The alteration is detected, the mismatch is clipped out and replaced with the correct nucleotide. Mismatch Repair Hmm. How does the enzyme know which is the strand with the error?? 5’AGGTCTAGATC3’ 3’TCTAGATCTAG5’ Mismatch Repair Adenine residues are methylated (CH3 group added) periodically The enzyme adenine methylase recognizes this sequence: 5’GATC3’ 3’CTAG5’ and adds the methyl group to each of the A residues: Mismatch Repair 5’GAmTC3’ 3’CTAmG5’ This is a stable modification that persists throughout the cell cycle. In the case of a mismatch, the repair enzyme uses this modification to differentiate between the strands. Mismatch Repair Newly replicated DNA is not yet methylated, so the repair enzyme chooses the methylated (old) strand as the template strand and fixes the error 5’AGGTCTAGAmTC3’ 3’TCTAGATCTAG5’ 5’AGGTCTAGAmTC3’ 3’TCCAGATCTAG5’ Double Strand Break Repair Ionizing radiation, in particular, causes breaks in both DNA strands. Also, several immune deficiency diseases and familial breast and ovarian cancers may be a result of double strand DNA breaks. Double Strand Break Repair Homologous recombinational repair: No template strand is available within the DNA strand (both are damaged), so the genetic information in the homologous sister chromatid is used. The undamaged region is recombined into the damaged region Usually occurs during late S/G2 phase of the cell cycle,. Double Strand Break Repair Nonhomologous recombinational repair (end joining): Does not involve homologous recombination Two protein complexes are involved in bringing damaged ends together and rejoining DNA strands.