Epigenetic Alterations in Cancer - Biomedical Informatics
advertisement

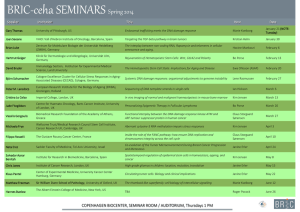
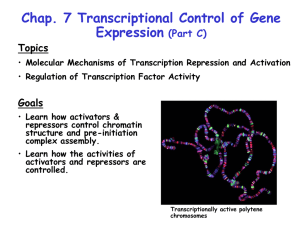
Chromatin Structure & Dynamics Victor Jin Department of Biomedical Informatics The Ohio State University Chromatin Walther Flemming first used the term Chromatin in 1882. At that time, Flemming assumed that within the nucleus there was some kind of a nuclear-scaffold. Chromatin is the complex of DNA and protein that makes up chromosomes. Chromatin structure: DNA wrapping around nucleosomes – a “beads on a string” structure. In non-dividing cells there are two types of chromatin: euchromatin and heterochromatin. Chromatin Fibers Chromatin as seen in the electron microscope. (source: Alberts et al., Molecular Biology of The Cell, 3rd Edition) 30 nm chromatin fiber 11 nm (beads) Nucleosome H2A H3 H2B H4 The basic repeating unit of chromatin. It is made up by five histone proteins: H2A, H2B, H3, H4 as core histones and H1 as a linker. It provides the lowest level of compaction of double-strand DNA into the cell nucleus. It often associates with transcription. 1974: Roger Kornberg discovers nucleosome who won Nobel Prize in 2006. Core Histones are highly conserved proteins - share a structural motif called a histone fold including three α helices connected by two loops and an N-terminal tail < 11 nm > Histone Octamer < 6 nm > Each core histone forms pairs as a dimer contains 3 regions of interaction with dsDNA; H3 and H4 further assemble tetramers. The histone octamer organizes 146 bp of DNA in 1.65 helical turn of DNA: 48 nm of DNA packaged in a disc of 6 x 11nm Nucleosome Assembly In Vitro 4 core histones + 1 naked DNA template at 4C at 2M salt concentration, from Dyer et al, Methods in Enzymology (2004), 375:23-44. 10,000 nm DNA compaction in a human cell nucleus 11 nm 30nm 1bp (0.3nm) DNA length compaction 10 m ball 12,000 Mbp 4 m DNA 400,000 x 1200 bp 400 nm DNA 35 x nucleus (human) 2 x 23 = 46 chromosomes chromatin fiber approx. 6 nucleosomes per ‘turn’ of 11 nm 30 nm diameter nucleosome disk 1.65 turn of DNA (146 bp) + linker DNA 6 x 11 nm 200 bp 66 nm DNA 6 - 11 x 0.33 x 1.1 nm 1 bp 0.33 nm DNA 1x base pair 92 DNA molecules compact size The N-terminal tails protrude from the core Histone Modifications Acetylation Me Ac Me Ub Su Methylation Ubiquitination Sumoylation P Phosphorylation ‘Histone Code’ Acetylation of Lysines Acetylation of the lysines at the N terminus of histones removes positive charges, thereby reducing the affinity between histones and DNA. This makes RNA polymerase and transcription factors easier to access the promoter region. Histone acetylation enhances transcription while histone deacetylation represses transcription. Methylation of Arginines and Lysines Arginine can be methylated to form mono-methyl, symmetrical di-methyl and asymmetrical dimethylarginine. Lysine can be methylated to form mono-methyl, di-methyl and tri-methylarginine. Methylation of Histone H3-K27 SUZ12 EED DNMT EZH2 PC K27 Functional Consequences of Histone Modification Establishing global chromatin environment, such as Euchromatin, Heterochromatin and Bivalent domains in embryonic stem cells (ESCs). Orchestration of DNA-based process transcription. Euchromatin A lightly packed form of chromatin; Gene-rich; At chromosome arms; Associated with active transcription. Heterochromatin A tightly packed form of chromatin; At centromeres and telomeres; Contains repetitious sequences; Gene-poor; Associated with repressed transcription. Bivalent Domains Poised state. The chromatin of embryonic stem cells has “bivalent” domains with marks of both gene activation and repression. In these domains, the tail of histone protein H3 has a methyl group attached to lysine 4 (K4) that is activating and a methyl group at lysine 27 (K27) that is repressive (above). This contradictory state may keep the genes silenced but poised to activate if needed. When the cell differentiates (right), only one tag or the other remains, depending on whether the gene is expressed or not. DNA Methylation N N N N O -O N O OH CH 3 DNA methyltransferase S-adenosylmethionine H deoxycytosine O -O N O OH H 5-methylcytosine CpG Islands CpG island: a cluster of CpG residues often found near gene promoters (at least 200 bp and with a GC percentage that is greater than 50% and with an observed/expected CpG ratio that is greater than 0.6). ~29,000 CpG islands in human genome (~60% of all genes are associated with CpG islands) Most CpG islands are unmethylated in normal cells. Chromatin modifications Mark Transcriptionally relevant sites Biological Role Methylated cytosine (meC) CpG islands Transcriptional Repression Acetylated lysine (Kac) H3 (9,14,18,56), H4 (5,8,13,16), H2A, H2B Transcriptional Activation Phosphorylated serine/threonine (S/Tph) H3 (3,10,28), H2A, H2B Transcriptional Activation Methylated argine (Rme) H3 (17,23), H4 (3) Transcriptional Activation Methylated lysine (Kme) H3 (4,36,79) H3 (9,27), H4 (20) Transcriptional Activation Transcriptional Repression Ubiquitylated lysine (Kub) H2B (123/120) H2A (119) Transcriptional Activation Transcriptional Repression Sumoylated lysine (Ksu) H2B (6/7), H2A (126) Transcriptional Repression Genome-wide Distribution Pattern of Histone Modification Associated with Transcription Source: Li et al. Cell (Review, 2007), 128:707-719 Li et al. Cell (review) 128, 707-719 Epigenetics Modifications of DNA (cytosine methylation) and proteins (histones) define the epigenetic profile. In 1942, Conrad Waddington first used “epigenetics” to describe the interactions between genome and environment that give rise to differences between cells during embryonic development. Currently, Epigenetics is the study of heritable changes in gene function that occur without a change in DNA sequence. Summarizes mechanisms and phenomena that affect the phenotype of a cell or an organism without affecting the genotype. Epigenomics is the study of these epigenetic changes on a genome-wide scale. Normal Cellular Functions Regulated by Epigenetic Mechanisms Correct organization of chromatin Genomic imprinting Silencing of repetitive elements X chromosome inactivation X-chromosome Inactivation 5 me 3’..pGpCp..5’ 5 me 5’..pCpGp..3’ transcriptional repressor MeCP2 co-repressor X-inactivation (also called lyonization) is a process by which one of the two copies of the X chromosome present in female mammals is inactivated. The inactive X chromosome is silenced by packaging in repressive heterochromatin. The choice of which X chromosome will be inactivated is random in higher mammals such as mice and humans. Once an X chromosome is inactivated it will remain inactive throughout the lifetime of the cell. Silencing initiated at Xic/XIC and spreads along chromososme. 5meC CpG DNA modification is observed in inactivated X chromosomes. 5meC binds transcriptional repressor MeCP2 (MethylC-binding Protein-2). MeCP2 binds Sin3 with RPD3 histone deacetylase. Sin3 Histone Deacetylase RPD3 Source: Jones et al. Nat.Genet. 19, 187 (1998) Epigenetic Diseases Some human disorders such as Angelman syndrome and Prader-Willi syndrom are associated with genomic imprinting. Involvement in cancer and development abnormalities. The emerging hypothesis of cancer stem cells (CSC). DNA Methylation and Gene Silencing in Cancer Cells CpG island CGCG CG Normal 1 MCGMCG MCG Cancer CG 2 3 MCG 1 3 MCG 4 CG CG 2 X MCG CG 4 C: cytosine mC: methylcytosine CG Progressive Alterations in DNA Methylation in Cancer Global + Hypomethylation Normal Region-Specific Hypermethylation Cancer Epigenetic Mediation of Gene Silencing DNMT Polycomb Repressors Histone-modifying Proteins Methyl-Binding Domain Proteins CpG Island Methylation: A Stable, Heritable and Positively Detectable Signal 1 2 3 4 Carcinoma 5 Normal Epithelia Dysplasia Carcinoma in situ Metastasis CpG Island Methylation: A Stable, Heritable and Positively Detectable Signal 1 2 3 4 Carcinoma 5 Normal Epithelia Dysplasia Carcinoma in situ Metastasis CpG Island Methylation: A Stable, Heritable and Positively Detectable Signal 1 2 3 4 Carcinoma 5 Normal Epithelia Dysplasia Carcinoma in situ Metastasis CpG Island Methylation: A Stable, Heritable and Positively Detectable Signal 1 2 3 4 Carcinoma 5 Normal Epithelia Dysplasia Carcinoma in situ Metastasis Epigenetic Alterations in Cancer Stem Cells Cancer Stem Cells: Stem cells arising through the malignant transformation of adult stem cells. Cancer Stem Cells Hypothesis: Cancer stem cells are the main driving force behind tumor proliferation and progression. Hallmarks of Cancer Stem Cells A cell residing in a tumor that – 1. has a capacity to remain in an undifferentiated state 2. has properties of asymmetric divisions and self-renewal 3. has metastatic and repopulation capacities at specific niches (microenvironment) in the body 4. gives rise to a tumor that is histologically identical to the one from which the CSC is derived The Evidence of Cancer Stem Cells First isolated from the patients of acute myeloid leukemia in 1997 by John Dick and colleagues at the University of Toronto. Isolated from two solid tumors, breast and brain cancers. ~1% cancer cells may be really cancer stem cells. More ChIP-chip Step 1: Rapid fixation of cells chemically cross-links DNA binding proteins to their genomic targets in vivo. Step 2: Cell lysis releases the DNA-protein complexes, and sonication fragments the DNA. Step 3: Immunoprecipitation (IP) purifies the protein-DNA fragments, with specificity dictated by antibody choice. Step 4: Hydrolysis reverses the cross-links within the released DNA fragments. Step 5: PCR amplification of ChIP DNA Step 6: PCR amplification on a known binding-site region for that protein will need to be performed using either conventional PCR methods followed by agarose gel electrophoresis or by quantitative PCR. Step 7: Labeling pool of protein-DNA fragments. Step 8: Hybridization of DNA onto microarrays featuring 60-mer oligonucleotide probes. Major types of array platforms NimbleGen Arrays: tiling arrays, promoter arrays, whole genome arrays. (http://www.nimblegen.com/products/chip/index.html) Agilent Arrays: promoter arrays, whole genome arrays. (http://www.chem.agilent.com/Scripts/Phome.asp) Affymetrix Arrays: tiling arrays, Chr21,22 arrays, whole genome arrays. (http://www.affymetrix.com/index.affx) Measurement of intensity of probes on the array The hybridized arrays were scanned on an Axon GenePix 4000B scanner (Axon Instruments Inc.) at wavelengths of 532 nm for control (Cy3), and 635 nm (Cy5) for each experimental sample. Data points were extracted from the scanned images using the NimbleScan 2.0 program (NimbleGen Systems, Inc.). Each pair of N probe signals was normalized by converting into a scaled log ratio using the following formula: •Si = Log2 (Cy5l(i) /Cy3(i)) Antibody Validation Confirming on a known target Different antibodies to same factor Antibodies to different family members siRNA-ChIP Antibodies to two components of a complex Antibodies to an enzyme/modification pair Confirming on a known target Comparison of biological replicates and antibodies to different E2Fs Loss of E2F6 ChIP signal after knockdown of E2F6 siRNA Reproducibility of promoter arrays using biological replicates •H3me3K27; Ntera2 cells •Top 1000 overlap •Top 1000 overlap •Promoter 1 •Promoter 2 Biological reproducibility on tiling arrays •500 kb region of chromosome 6 •500 kb region of chromosome 1 Amount of Sample Per ChIP Number of cells Chromatin input ChIP output 1x107 200 µg 150 ng 1x106 20 µg 10 ng 5x105 10 µg 1.3 ng 1x105 2 µg 300 pg 1x104 200 ng 30 pg Amount of Sample Per ChIP Number of cells Chromatin input ChIP output 1x107 200 µg 150 ng 1x106 20 µg 10 ng 5x105 10 µg 1.3 ng 1x105 2 µg 300 pg 1x104 200 ng 30 pg Miniaturization •Standard ChIP Protocol (1x107 cells; WGA2) • Promoter Arrays • Genome Tiling Arrays •MicroChIP Protocol (10,000-100,000 cells; WGA4) • Promoter Arrays • Genome Tiling Arrays Reproducibility of MicroChIP Protocol