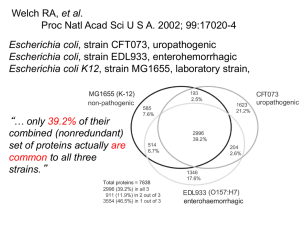
Plant genome
advertisement
Plant molecular genetics • • • • • • • • • • • Plant genome Chromatine and DNA methylation RNA interference Genome of plastids and mitochondria Transposible elements Viruses Classical genetic mapping Transgenosis and reverse genetics Genomics, next generation sequencing Transcriptomics Proteomics Components of plant genome • nuclear genome = genome sensu stricto • plastids - plastome • mitochondria - chondriome Plant genome sizes 54 Mbp – Cardamine amara 124 852 Mbp - Fritillaria 149 000 Mbp - Paris japonica - currently the largest (not only plant) http://data.kew.org/cvalues/ Plant genome sizes 10 Mb Ostreococcus (single cell alga) 54 Mb Cardamine amara 64 Mb Genlisea aurea Ratio of globe volumes differing 3000 times 125 Mb Arabidopsis 500 Mb Oryza 5 000 Mb Hordeum 17 000 Mb Triticum 84 000 Mb Fritillaria (largest diploid) 143 000 Mb Paris (oktaploid) - Angiosperms – size differences up to almost 3 000 times - Gymnosperms – genome sizes often around 10 000 Mb - Gene number differences much lower (approx. 20 – 200 fold) Plant genome sizes What we can deduce? - Genomes are increasing in evolution - Average increase is higher in Monocots C-value paradox - there is no strong correlation between complexity of an organism and the size of its genome • C-value = size of genome in non-replicated gamete genome size (bp) = (0.910 x 109) x DNA content (pg) DNA content (pg) = genome size (bp) / (0.910 x 109) 1 pg = cca 910 Mbp; MW (1 bp) = cca 660 Da • genomes of related organisms often strongly differ in size causes: - duplications of whole genomes (polyploidization) or chromosome segments - replication of invasive DNA (transposable elements) - but reductions also possible (recombination – diploid cotton sp.) Sequences in plant genomes Unique sequences – genes, but also non-coding (!) Repetitive: • Duplications of chromosomal regions • Medium repetitive DNA – Tandem repeats of rRNA, tRNA a histon genes – Gene families with multiple members – Transposable elements – also high repetitive • Highly repetitive – low complexity DNA - Tandem arranged simple sequence repeats (SSR) – Centromers (180 bp repeat Arabidopsis) a telomers (TTTAGGG)n Types of sequences in plant genomes • Unique sequences – coding genes, but also noncoding regulatory (!) • Medium repetitive DNA – Tandem repeats of rRNA, tRNA a histon genes – Gene families with multiple members – Transposable elements – also highly repetitive • Low complexity DNA (highly repetitive) – Tandem arranged simple sequence repeats (SSR) – Centromers (180 bp repeat Arabidopsis) a telomers (TTTAGGG)n - some behave as satelite DNA Aside – term definition: sequence complexity (~ the amount of information) repetitive AAAAAAAAAAAAAAAAAAAAA complexity 1 (21xA) ATCATCATCATCATCATCATC complexity 3 (7xATC) (what is the complexity if it is a coding sequence?) unique ATCGTATCGCGATTTTAACGT complexity 21 (1xAT…) - unique x repetitive – depends on the size of the evaluated frame (= size of analyzed DNA fragments) Sequence complexity of plant genomes Higly repetitive Medium repetitive Unique Sequence complexity Examples of repetitive DNA representation in u Soybean and Silene (clusters of related sequences) Silene latifolia Gypsy, copia = retrotransposon families clDNA = chloroplast DNA (partially contamination, but also recent insertions) Measuring of genome complexity reasociation kinetics • DNA fragmented to 300 - 500 bp, denatured • Monitoring of reassociation in time - separation (chromatographic) of ss and ds DNA • Analysis of kinetics (Cot curves) shows representation of various types of repetitive DNA – rare sequences reasociate more slowly that repetitive Reasociation kinetics depends on sequence complexity Eucaryotic genomes usually contain three fractions of sequences with different complexity Low complexity = highly repetitive Middle repetitive Unique sequences = High complexity Reasociation kinetics of small and large genomes Unique Medium repetitive Highly repetitive Repetitive sequences can be easily detected in situ FISH = fluorescent in situ hybridization (possible even with unique seq.) 180 bp A.th. 45S rDNA Crocus copia A.th. tandem repeats dp5a1 wheat (Heslop-Harrison, Plant Cell 12:617, 2000) Subtelomeric repeats in rye (Heslop-Harrison, Plant Cell 12:617, 2000) Telomers in rye (TTTAGGG)n Differences in small and large genome arrangements large genomes: genes present in „gene-rich islands“ isolated with long regions of repetitive DNA Reconstruction of gradual cummulation of transposable elements in maize genome In Panicum in the presented region no transposible elements, in maize 60 % of its size Plant Genome Sequencing http://genomevolution.org/wiki/index.php/Sequenced_plant_genomes April 13 – less complete in gray Large Genome Sequencing - sequencing per partes (separated chromosomes) - sequencing of non-methylated DNA (= transcriptionally active) - sequencing of ESTs Aside – term definition: Expressed Sequence Tags (ESTs) - short sequenced regions of cDNA (300-600 nt) - mostly gene segments (primarily from mRNA) - alternative sourse of coding sequences for large genomes (rapid and inexpensive) Weak points: - highly redundant, incomplete (!) - problems: various transcript levels - gene expression regulated spatially and temporally, developmentally, environmentally - regulatory sequences not represented (promotors, introns,...) Expressed Sequence Tags (ESTs) Preparation of EST library - mRNA - RT with oligoT primer cDNA -cleavage of RNA from heteroduplex RNAseH - 2nd strand cDNA synthesis - cleavage with restriction endonuclease - adaptor ligation cloning sequencing Aside: Arabidopsis thaliana 1 week 3 weeks the most important model of plant biology 4 weeks 6 weeks Arabidopsis genome: 125 Mbp genes ESTs TEs genes ESTs TEs genes ESTs TEs genes ESTs TEs genes ESTs TEs High density low density Total gene number prediction in time (after whole genome sequencing) Genome of Arabidopsis statistics Value Feature DNA molecule Chr.1 Length (bp) Top arm (bp) Bottom arm (bp) 29,105,111 14,449,213 14,655,898 Base composition (%GC) Overall Coding Non-coding Number of genes Gene density (kb per gene ) Average gene Length (bp) Average peptide Length (bp) Exons Number Total length (bp) Average per gene Average size (bp) Number of genes With ESTs (%) Number of ESTs Chr.2 Chr.3 Chr.4 Chr.5 SUM 19,646,945 3,607,091 16,039,854 23,172,617 13,590,268 9,582,349 17,549,867 3,052,108 14,497,759 25,53,409 11,132,192 14,803,217 115,409,949 33.4 44.0 32.4 35.5 44.0 32.9 35.4 44.3 33.0 35.5 44.1 32.8 34.5 44.1 32.5 6,543 4.0 4,036 4.9 5,220 4.5 3,825 4.6 5,874 4.4 2,078 1,949 1,925 2,138 1,974 446 421 424 448 429 35,482 8,772,559 5.4 247 19,631 5,100,288 4.9 259 26,570 6,654,507 5.1 250 20,073 5,150,883 5.2 256 31,226 7,571,013 5.3 242 60.8 56.9 59.8 61.4 61.4 30,522 14,989 20,732 16,605 22,885 25,498 132,982 33,249,250 105,773 + hundreds of MIR genes - role in regulation of gene expression Gene function The majority of plant genes form gene families Number of paraloques • gene families are often in tandem arrangement, but also spead in the genome • tandem repeats are composed of near, but also far paralogues (recombinations) • duplications of long chromosomal regions Aside – terms definition: Homologous genes genes with similar sequences derived from the same ancestral gene (quantification – sequence identity, similarity) • Paralogous genes genes with similar sequences derived from the same ancestral gene present at different loci within the same genome. • Orthologous genes genes in different species that are similar to each other because they originated from a common ancestral gene in a common ancestor. (if more paralogues are present – genes serving the same function are regarded to be orthologs) Orthologues vs. paralogues Orthologous genes Species A Gene A” Ancestral Gene A Species Species B Gene A’ Paralogous genes = genes duplicated within the species Species A Ancestral Gene A Species Gene A” Gene A’” Paralogous genes Species B Gene A’ Mechanisms of gene duplications (increase in paralogue number) • tandem duplication • transpozition • segmental duplications • whole genome duplications Differences in genes/gene families in genomes Genes Gene families Arabidopsis x Populus – large overlap, about 1,5 times more paralogues in poplar (Arabidopsis + Populus) x Oryza – many genes specific for Monocots Arabidopsis is ancient tetraploid (as well as probably the majority of plants) Duplicated chromosomal regions form about 60 % of genome (67.9 Mb) Polyploidization significantly increases genome (and organism) plasticity and played very important role in plant (genome) evolution; About 30-80% plant species are polyploid Polyploidization in Angiosperm evolution Fawcett et al. 2009 Dating of whole genome duplication according to the number of synonymous mutations per synonymous site - Ks Ks=3/2,66 Phe Leu Met Val UUU CUA AUG GUU UUC UUG AUG GUU 0 0 1/3 1/3 0 1 0 0 0 0 0 1 Gene number = number of syn. sites Comparisons of paralogue pairs Peaks indicate genome duplications Ks Fawcet et al. 2013 Polyploidization in plant evolution • 35 % species neopolyploids • most species repeatedly polyploid in evolution • viable aneuploid variants – (frequetly after allopolyploidization – hexaploid wheat) stabile wheat lines with missing chromosomal arm (of homeologic chromosome) Blue dots – duplications, asterix – triplication K-T (Fawcett et al. 2013) Polyploidization - fusion of non-reduced gametes or endoreduplication n=x=4 n=x=4 n=x=4 x x 2n = 4x = 16 n=x=7 Spontaneous duplication (endoreduplication) 2n = 4x = 22 autopolyploidy allopolyploidy Similar frequency in polyploidic plant species Chromosome doubling is necessary for meiosis in hybrids species A species B X sterile Genome duplication fertile Preferential pairing of homologous chromosomes Related from different species (homeologous) can also pair Allopolyploidic genomes in Brassica genus BB Species Caryotype Genom e Brassica rapa 2n = 2x = 20 A B. nigra 2n = 2x = 16 B B. oleracea 2n = 2x = 18 C B. juncea 2n = 4x = 36 AB B. napus 2n = 4x = 38 AC B. carinata 2n = 4x = 34 BC Brassica nigra BBCC AABB Brassica juncea Brassica carinata CC Brassica olarecea Ancient interspecies hybrids AACC Brassica napus AA Brassica rapa Allopolyploid tobacco species – DNA size changes Fade of duplicated genes differ (gene dosage balance theory) genes encoding interacting proteins “connected genes“ (signal pathways, complex subunits, …) easily preserve in genome after duplication • - loss or partial duplication of one component results in gene inbalance decreasing fitness, - whole duplicated complex can be specialized for a new function and increase organism complexity -secondary function probably present already in the ancestral complex (pathway), but only duplication allowed adaptive evolution for both functions without selection constrains - Escape from adaptive conflict - EAC model • other „single genes“ more easily lost after genome duplication, but can be preserved after individual duplication - most of duplicated genes is lost after whole genome duplication - loss is not as even (↑) in both copies - probably frequent epigenetic marks in one copy (methylation) - preferential gene loss and mutagenesis of methylated copy - gene conversion and homogenization can occur (!) de novo allopolyploids (~ rape seed) – recombinations preferentially in homeologous chromosomes without preference of any parental genome (= homologní, v jednom genomu, ale původem od různých rodičů) Changes in newly formed allopolyploid genome: - DNA methylation changes - losses of parts or whole chromosomes (aneuploidy – decreased fertility) - frequent activation of TE - expression of homeologous genes is not usually additive - transcriptome usually more reduced than genome - different regulation of expression - often organ specific expression of genes from each parent, new sites of expression, new regulation - „divergent resolution“ - speciation (different gene loss in individuals - lethality in F2, - absence of essential gene = reproduction barrier Plants can survive also with haploi genome! - reprogramming of male or female gametophyte development in vitro – no gamete formation, but development resembling embryogenesis - usually from immature microspores = androgenesis - female gametophyte = gynogenesis - haploid plants are sterile - through endoreduplication (colchicin or spontaneous) – completely homozygous plants – dihaploids Androgenesis in rape seed (pollen embryogenesis) ... But genomes are still similar Colinearity, syntheny Paterson et al., Plant Cell 12: 1523-1539, 2000 „Syntheny“ is usually missused to describe colinearity Syntheny = orthologous loci in two species on the same chromosome Species A Ancestral Species A B C Species B A’ C’ B’ C” B” A” Colinearity = group of loci in two species on a chromosom in the same order Species A Ancestral Species A B C Species B A’ B’ C’ A” B” C” Changes in colinearity caused by chromosomal arm inversion Colinearity of Poaceae genomes Colinear regions differ mainly in repetitive DNA Summary: • Current plant genomes result from repeated cycles of partial and complete duplications, followed by reduction and modification of duplicated sequences. • There are no genomes without redundancy. • Plant genomes are still very dynamic. • High portion of genome consists of repetitive DNA