LysM-type Mycorrhizal Receptor Recruited for Rhizobium Symbiosis
advertisement
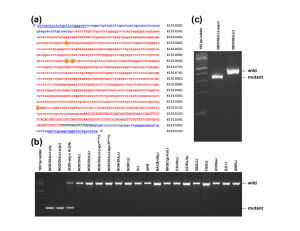
Rik Op den Camp, Arend Streng, Stephanie De Mita, Quingpin Cao, Elisa Polone, Wei Liu, Jetty S. S. Ammiraju, Dave Kudrna, Rod Wing, Andreas Untergasser, Ton Bisseling, Rene Geurts Rik Op den Camp ◦ PhD student Molecular Biology at Wageningen University Arend Streng ◦ PhD researcher at Wageningen University STEPHANE DE MITA ◦ Ph.D, molecular evolutionist ◦ Do mutually beneficial symbioses cause strong selective pressures and represent an evolutionary driving force comparable to hostpathogen relationships? Focused on the independent evolution of the recognition of the rhizobial signal that starts the symbiotic interaction...The nod factor Identify the genetic constraints which caused such Nod Factor signaling ◦ Compare it to the Myc Factor Soil bacteria that help to fix nitrogen after entering into the root nodule of a plant cell. Usually unique for legumes ◦ However, it has been found that Parasponia, a non legume has a rhizobia nodule symbiosis. ***Symbiosis is induced for all by specific Nod factors Plant ◦ nitrogen in a usable form because of nitrogen fixation Rhizobia ◦ Reduced carbon ◦ Protective environment Legume Non legume Parasponia Key: Both have symbiosis with bacteria to fix nitrogen. Fabaceae -Lipochitooligosaccharides -Nod factors are perceived by specific Lys M receptor kinases -Activate a signaling cascade -Results in the introduction of a dominant active form of calcium/calmodulindependent kinase (CCaMK) Allow for rhizobia to enter plant cell Signal to allow for the response of a plant cell to be penetrated by a fungus Induces a Calcium spike Part of symbiosis between plant and fungi Lipochito-oligosaccharides Myc Factor receptors, like Nod factor receptors are presumed to activate a common pathway to initiate this symbiosis. Thus, before these to differentiated maybe a DUAL PURPOSE RECEPTOR existed! This experiment sought to prove, if not find its existence. Parasponia andersonii = a plant member of the non-legume, nitrogen fixing, group Sinorhizobium = a broadly occurring bacteria used to illustrate Nod factor use Medicago truncatula MtCCaMK= a dominant active kinase that spontaneously forms nodules in both legumes and Parasponia They introduced a normal strain of Sinorhizobium to P. andersonii and a mutant strain to a different batch of P. andersonii. The Mutant bacteria had the Nod factor disabled and NO nodules formed. The Normal bacteria produced healthy, nitrogen fixing nodules. By using an extremely dominant and fast acting kinase, Medicago truncatula MtCCaMK, nodules were able to spontaneously form on both the legume and P. andersonii. -There is a common signaling pathway used between two organisms in order to gain functioning nodules - This pathway is activated upon a Nod Factor Perception - Legumes have two factors to initiate the common pathway, we hope to find that P. andersonii only has one! -The only one capable of the mechanisms needed is the MtNFP/LjNFR5 factor What does this mean? They need to find the gene(s) that code for this and compare it to relative species in order to varify its unique properties Method to Identify Gene: Southern Blotting and Sequencing (Very Similar to PCR) MtNFP/LjNFR5- like receptor named PaNFP Fulfills Dual Function of intracellular lifestyle: both the Arbuscular Mycorrhiza and rhizobium. Unique to Parasponia as only having this one receptor! Favor: Mycorrhizal signaling pathway including the Myc receptor being recruited by the legumes to make a common signaling pathway Nod factors then have emerged then by gene duplication Support: Chitooligosaccharides which is the backbone of the Nod factor is a fungal characteristic Strength ◦ Detailed evidence to prove their interpretation of the data Weakness ◦ Does not disprove the second way in which states the recognition of the Nod factor was created without the Myc factor involved How does a plant then clarify whether it is a fungi or a bacteria making the symbiosis? ◦ Look at the differences in the spike of Calcium to see if large enough difference can verify the symbiosis.


![Symbiosis[1]](http://s2.studylib.net/store/data/005449742_1-2c9de7b7b178f521480e9109673f342e-300x300.png)







