Chapter 4 Phylogenetics I
advertisement

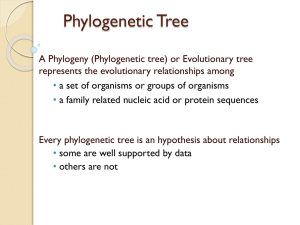
Read Chapter 4 All living organisms are related to each other having descended from common ancestors. Understanding the evolutionary relationships between groups enables us to reconstruct the tree of life and gain insight into history of evolutionary change. Phylogeny is the study of the branching relationships between populations over evolutionary time. A phylogenetic tree is built up by analyzing the distribution of traits across populations. Figure 4.2 Phylogenies at different scales Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company A trait (or character) is any observable characteristic of an organism. Could be anatomical features, behaviors, gene sequences, etc. Traits are used to infer patterns of ancestry and descent among populations. These patterns are then depicted in phylogenetic trees. By mapping other traits onto trees it is possible to study the sequence and timing (history) of evolutionary events. Figure 4.4 Traits and trees Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company It’s important to bear in mind that phylogenetic trees are hypotheses about the evolutionary relationships between groups. When additional evidence is acquired it can be used to test a tree. Each branch tip represents a taxon (a group of related organisms). Interior nodes (where branches meet) represent ancestral populations that are the common ancestors of the taxa at the ends of the branches. Figure 4.6 Interior nodes represent common ancestors Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Phylogenetic trees are generally drawn in either a Tree format or a Ladder format. They convey the same information about the relatedness of taxa Figure 4.5 Two equivalent ways of drawing a phylogeny Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company It is important to remember that a particular set of evolutionary relationships can be depicted in multiple different ways in a phylogenetic tree. Any node in a phylogenetic tree can be rotated without altering the relationships between taxa. Figure 4.7 Rotating around any node leaves a phylogeny unchanged Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Figure 4.8 Rotating phylogenetic trees Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company The purpose of building phylogenetic trees is to use them to figure out the evolutionary relationships between taxa and to identify “natural” groupings among taxa, those that reflect their true evolutionary relationships. A key idea is that natural groupings called clades are monophyletic groups. Clade: a group of taxa that share a common ancestor. Monophyletic group: consists of an ancestor and all of the taxa that are descendants of that ancestor. In the next slides elephants, manatees and hyraxes plus their common ancestor form a monophyletic group. Similarly tapirs, rhinoceroses and horses plus their common ancestor form another monophyletic group. Figure 4.11 Monophyletic clades of mammals Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company A taxon is polyphyletic if it does not contain the most recent common ancestor of all members of the group. A polyphyletic group requires the group members to have each had an independent evolutionary origin of some diagnostic feature. E.g. Referring to Elephants, rhinoceroses and hippopotamuses as “pachyderms.” Pachyderms are a polyphyletic group because each group evolved thick skin separately. Elephants, rhinos and hippos would form polyphyletic group Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company A taxon is paraphyletic if it includes the most recent common ancestor of a group and some but not all of its descendents. An example of a paraphyletic group among vertebrates would be “fish.” All of the tetrapods (four-legged animals) are descended from lobefinned fish ancestors, but are not considered “fish” hence “fish” is a paraphyletic group because the tetrapods are excluded. Figure 4.12 Phylogenetic tree of the vertebrates Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Trees we’ve seen so far have been rooted and these trees give a clear indication of the direction of time. However, computer programs that produce phylogenetic trees often produce unrooted trees. In an unrooted tree branch tips are more recent than interior nodes, but you cannot tell which of multiple interior nodes is more recent than others. Figure 4.13 Unrooted tree of proteobacteria Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company An unrooted tree can be rooted at any point and depending where it is rooted very different rooted trees will be produced. Figure 4.14 Rooted trees from unrooted trees Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Obviously, there is only one true tree of evolutionary relationships and we would like to identify that tree. To do that we need to root the tree correctly. One of the easiest ways to root a tree is to use an outgroup to root it. An outgroup is a close relative of the members of the ingroup (the various species being studied) that provides a basis for comparison with the others. The outgroup lets us know if a character state within the ingroup is ancestral or not. If the outgroup and some of the ingroup possess a character state then that character state is considered ancestral. Consider an unrooted tree of four magpie species. To root the tree we need a group that split off earlier from the lineage that led to these four species of magpies. Azure-winged magpie is a suitable outgroup. One this is added to the unrooted tree we can root the tree. In some phylogenetic trees branches are drawn with different lengths. In these trees the branch lengths represent the amount of evolutionary change that has occurred in that lineage. Figure 4.15 Cladograms and phylograms Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Phylogenetic trees can be used to generate hypotheses about the evolution of traits. This is done by mapping the trait states on the tree and trying to reconstruct the simplest (most parsimonious) explanation that accounts for the observed distribution of traits. Light sensitive pigments called opsins are responsible for color vision. Humans have three different cone opsins. Other vertebrates have as many as four or as few as two opsins. By mapping the presence or absence of different opsins onto a phylogenetic tree of vertebrates we can attempt to reconstruct the evolutionary history of color vision in these vertebrates. Figure 4.20 Evolution of tetrapod visual opsins Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company It is clear that the ancestral trait is to possess four opsins (as both birds and reptiles do). The mammal lineage appears to have lost two opsins (probably because the animals were nocturnal) and one opsin was later re-evolved on the lineage leading to old world primates and humans. Homologous traits are those derived from a common ancestor. E.g. all mammals possess hair. This is a homologous trait all mammals share because they inherited it from a common ancestor. Analagous traits are shared by different species not because they were inherited from a common ancestor but because they evolved independently. Figure 4.21 Homologous and analogous traits Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Divergent evolution occurs when closely related populations diverge from each other because selection operates differently on them. Such new species will possess many homologous traits in common. Analagous traits are the result of a process of convergent evolution whereby the same or similar solution to an evolutionary problem is converged upon by different organisms independently of each other. Figure 4.22 Convergent evolution for coloration Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Figure 4.23 Convergent evolution in body forms Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company When building a phylogenetic tree we want to use characters inherited from ancestors. Such a character found in two or more taxa is referred to as a shared derived character or synapomorphy. Example B on the next slide is a synapomorphy. Figure 4.24 Derived traits Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company If all shared traits were shared derived traits tree-building would be straightforward. However, many traits are not e.g. analagous traits We want to avoid including analagous traits when constructing phylogenetic trees because they can mislead us. An analagous trait in trees is referred to as a homoplasy. Figure 4.25 An example of homoplasy Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Another way in which we could be mistaken is if a new trait arises in a lineage and is not shared with other taxa. This is called a symplesiomorphy. In the next slide light coloration has recently arisen in taxon 3. If we thought dark coloration was a shared derived character we would group species 1+2, but it isn’t it is an ancestral trait. Figure 4.26 Derived traits and symplesiomorphy Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Several strategies exist to limit homoplasies and synapomorphies. 1. use traits that change relatively slowly in evolutionary time 2. use many traits to build the tree 3. use multiple outgroups to help identify ancestral values of traits. In next slide relationships between species 1,2,3 are unclear [this branching arrangement is called a polytomy] Species 1 and 2 are dark and 3 is light. Species 01 and 02 are outgroups and two cases (01 and 02 both dark and 01 and 02 both light) are shown. Figure 4.27 Using outgroups to infer the ancestral state Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Scenario A requires only a single evolutionary change. Scenarios B and C require two changes each. Figure 4.28 Case 1: The outgroups help resolve the polytomy Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Scenario A and scenario B are equally likely so in this case outgroup does not help resolve the polytomy. Figure 4.29 Case 2: The outgroups do not help resolve the polytomy Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company In next slide two characters with ancestral states of A and C [outgroups possess these states]. Each of the two characters is used to resolve a polytomy. Figure 4.30 Synapomorphies at different levels Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Phylogenetic trees are hypotheses about evolutionary relationships among groups . When traits are mapped onto a phylogeny the distribution of traits can also be used to generate hypotheses about the evolution of those traits. A large number of very venomous snakes possess highly developed venom delivery systems that include large grooved or hollow fangs, large venom glands, and, of course, venom. Figure 4.31 Snake fangs and venom Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Snakes with such delivery systems belong to the families: Viperidae (e.g. vipers and rattlesnakes), the Elapidae (e.g. cobras and mambas) and include some members of the Atractaspididae (asps). Cobra http://img.gawkerassets.com/img/1 87dj0ekcb9kijpg/original.jpg Gaboon Viper http://pphotographyblog.blogspot.com/2011/11/veno mous-gaboon-viper.html For many years it was considered that advanced venom delivery systems had evolved independently in each family and thus were analagous traits. Researchers had assumed there would be no venom without a delivery system. More recent phylogenetic analysis and more morphological studies however have led to this idea being reevaluated. For example, specialized oral secretory grooves are found in many snakes and numerous snakes have been discovered to be able to produce salivary toxins in a gland called Duvernoy’s gland. Figure 4.32 Phylogeny of advanced snakes (Caenophidia) Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Suspecting that if basic toxin production capacity is widespread in snakes Bryan Fry a herpetologist thought that toxin production might be homologous in snakes and had arisen early in the group. If this was true, there might be many snakes capable of producing toxic venom among those that lacked a sophisticated delivery system. When Fry examined the salivary secretions of the supposedly non venomous rat snake he discovered that the commonest peptide in the saliva was a close homologue of the threefinger toxins (3FTX), potent neurotoxins, that elapids produce. Having discovered venom in supposedly non-venomous snakes Fry wondered if snake venom might be homologous with the venom found in Gila Monsters (a lizard) which would imply venom had been inherited from the common ancestor of both. If venom evolved early, other lizard descendents of the common ancestor of snakes and gila monsters might also produce venom. Fry produced a phylogenetic tree to identify such potentially venomous lizards Figure 4.33 Venomousness as a homologous trait between snakes and Gila monsters Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Fry discovered nine genes for toxins that were shared between snakes and lizards. Also found an Australian lizard that produces a toxin otherwise found only in rattlesnake venom and that some monitor lizards produce a toxin that reduces blood clotting and greatly lowers blood pressure. The significance of Fry’s work is that it shows how the mapping of traits on phylogentic trees may lead to unexpected research directions and reveal hidden characteristics of organisms. Vestigial traits (those with no known current function but that were important in the evolutionary past) are often useful in constructing phylogenetic trees. For example, the nictitating membrane in birds and the plica semilunaris indicate common descent from an ancestor that had a nictitating membrane that became vestigial in mammals. Figure 4.36 The nictitating membrane Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Vestigial traits provide a test of Darwin’s theory of evolution by common descent. If organisms evolved from common ancestors via a branching process we would expect to see vestigial traits only in those organisms that share a common ancestor that existed after the trait evolved. For example we would expect perhaps to find vestigial limbs in some vertebrates, but not in fish or annelid worms. Figure 4.38 Common ancestry predicts where we should find vestigial limbs Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Find the most recent common ancestor of species 3,5 and 6 Review Question 4.1 Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company What would this tree look like if it were rotated around (i) Node A (ii) Node B (iii) both nodes A + B? Review Question 4.3 Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company (i) 1, 3, 2, 4, 5, 6 (ii) 1, 6, 5, 4, 3, 2 (iii) 1, 6, 5, 4, 2, 3 Which pairs of species are more closely related? (i) 4&5 or 5&7? (ii) 1&2 or 2&7? (iii) 3&5 or 2&4? Review Question 4.5 Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company (i) 5&7 (ii) 2&7 (iii) 3&5 According to the diagram which of these five fraits do (i) sharks (ii) turtles have? Review Question 4.10 Evolution, 1st Edition Copyright © 2012 W.W. Norton & Company Shark : jaws Turtle: jaws, dentary bone, lungs. Do the other problems in your text!