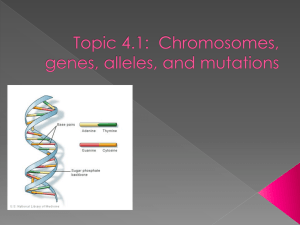
chapter eight:
microbial genetics
the hereditary material
Griffith 1927 & Avery, et al. 1944
the “transforming principle” coined by Griffith, identified by Avery
the hereditary material
Hershey Chase, 1952
the bacterial chromosome
plasmids
• F factor (conjugative plasmid)
• dissimilation plasmids
• R factors
horizontal & vertical gene transfer
antiparallel replication
vertical gene transfer (VGT): DNA replication
synthesis requires primers & the 3΄ OH
horizontal gene transfer (HGT): gene expression
simultaneous transcription & translation
HGT: recombination
RecA & chromosomal recombination
insertion sequences & jumping genes
recombination: transformation
recombination: transduction
recombination: conjugation
genetic transfer
Transfer
Effects
Transformation
demo
naked/free DNA from donor
DNA binding proteins on recipient
RecA needed for DNA fragments
transposons chromosome
plasmids self-contained
Transduction
(specialized)
Phage incorporates bacterial donor DNA, delivers to recipient
Conjugation
F+ cells
F- cells
Hfr cells
F factor codes for sex pilus, delivers donor DNA
Contain F factor (donor cell)
Lack F factor (recipient cell)
High frequency of recombination (donor cell)
F factor integrated into donor chromosome at integration
point, donates partial F factor from point of transfer and
chromosome portion to recipient cell.
Recombined F- cell
F+ and F- F+ and F+
Hfr and F- Hfr and recombinant F-
regulating bacterial gene expression:
constitutive enzymes operons
regulating gene expression*
* decreased levels of cellular glucose create high cAMP levels which
further regulate the expression of lactose catabolizing enzymes- this
will not be discussed in this class
transcriptional control
inducible operon:
effector effects by inhibiting repressor
= inducer
repressible operon:
effector effects by activating repressor
= corepressor
quorum sensing & gene regulation
• B. subtilis sporulation
– cell density = CSF & ComX ComS competence
– cell density & CSF = ComS inhibited sporulation
• Gram negative biofilm formation
– acylated homoserine lactones
(HSLs) in loss of flagella
– sessile microbes initiate biofilm
formation
• P. aeruginosa virulence
– high cell density activates virulence genes disease
Chapter Eight Learning Objectives
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
What did the work of Griffith, Avery and Hershey & Chase contribute to the field of
biology?
How is the bacterial chromosome different from the eukaryotic chromosome? What
other molecule contains useful genetic information for prokaryotes? Compare and
contrast DNA replication in eukaryotes vs. prokaryotes.
Why does the replication of every DNA molecule start with a short segment of RNA?
Define: vertical gene transfer, horizontal gene transfer, DNA replication, gene expression,
transcription, translation, conjugation, transduction and transformation.
How is gene expression in prokaryotes different from eukaryotes, both in the timing of
transcription & translation and in how transcription is regulated?
How do the RecA protein and transposons enable novel DNA to be integrated and used in
the recipient cell? Discuss this for both transformation and transduction.
Define F factor, F+ cell, F- cell and Hfr cell. Understand what happens when F+ , F- & Hfr
cells interact during conjugation.
Describe the mechanisms of inducible and repressible operons. Include the role of
promoters, operators, effectors, inducers, repressors and co-repressors in your answer.
Discuss the levels of bacterial control of gene expression, paying particular attention to
post-translational and transcriptional control, as discussed in lecture.
What is quorum sensing? How does it relate to gene expression, particularly as relates to
sporulation, biofilm formation, competence and virulence genes.
chapter nine:
biotechnology
biotechnology and recombinant DNA
• biotechnology: using recombinant DNA (rDNA) cells
– using vectors to produce clones
• therapeutic applications
– human enzymes and other proteins
– subunit vaccines
– viral DNA vaccines
– gene therapy
– disease ID
• mutant screening!!!
– natural or mutagen-induced
• >2000 Abx compounds
• penicillin 1000× stronger than wild type
– cloned & expressed recombinant DNA technology
rDNA technology
pharmaceutical products
restriction endonucleases
in vivo: defense system, cut only non-methylated DNA
in vitro: molecular scissors
making RFLPs: restriction endonucleases
making & moving rDNA: plasmid vectors
shuttle vectors
finding rDNA: blue/white colony selection
pBluescript™ vectors
moving rDNA: viral vectors
pathogen detection: PCR (second animation)
E. coli O157:H7 outbreak
chapter eight:
microbial genetics
mutation frequency
• change in the genetic material
• spontaneous
– no mutagen
– 109 per bp
– 106 genes
• mutagens freq.
105 – 103 per gene
mutation types
• base substitution (point mutation)
– silent
• 3rd G to any other base = glycine (redundancy)
– protein change
• missense, nonsense, frameshift mutation
mutagens
mutation repair
• photolyase repair
– separate thymine dimers
• nucleotide excision repair
– various damage repaired
– UvrA, UvrB, UvrC, UvrD
(DNA helicase)
• SOS recA repair
– cell cycle arrested
– DNA repair & mutagenesis induced
replica plating: negative mutant selection
wildtype auxotroph mutants die
the Ames test: positive mutant selection &
carcinogen identification
auxotroph wildtype mutants grow
Chapter Nine & Eight B Learning Objectives
CHAPTER 9
1. Define biotechnology & recombinant DNA technology. What applications were discussed in lecture
which utilize this technology?
2. Discuss how recombinant DNA molecules are made using restriction enzymes. What are the steps used
in making these recombinant molecules? How do both plasmids & viruses play a role in expressing
recombinant DNA molecules?
3. There are four essential regions on a shuttle vector. What are they, and what do they do? How do they
help to identify in vitro transformed cells?
4. Describe the process of PCR to amplify a DNA template. How can thistechnologies be used to identify a
microbial pathogen?
CHAPTER 8B
1. Define: silent, missense, nonsense and frameshift mutation. How can these errors be repaired in a cell?
2. How does the term auxotroph relate to mutant selection?
3. Why is replica plating necessary for the indirect selection of mutants?
4. What is the Ames test? How and why does it result in positive mutant selection?