Chapter 7
GENES AND PROTEIN SYNTHESIS
ONE GENE-ONE POLYPEPTIDE HYPOTHESIS
DNA contains all of our
hereditary information
Genes are located in our
DNA
~25,000 genes in our
DNA (46 chromosomes)
Each Gene codes for a
specific polypeptide
MAIN IDEA
Central Dogma
Francis
Crick (1956)
OVERALL
PROCESS
Gene 1
Gene 3
DNA molecule
Transcription
DNA
Gene 2
to RNA
Translation
of
amino acids
into
polypeptide
Using RNA
DNA strand
Assembly
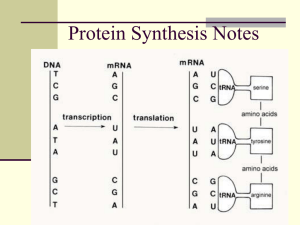
TRANSCRIPTION
RNA
Codon
TRANSLATION
Polypeptide
Amino acid
KEY TERMS
RNA transcription
Initiation, Elongation, Termination
TATA box
Introns, Exons
mRNA, tRNA, rRNA
Translation
Ribosome
Codon
Amino Acids
Polypeptide
DNA
RNA
Double stranded
Single stranded
Adenine pairs with Thymine
Adenine pairs with Uracil
Guanine pairs with Cytosine
Guanine pairs with Cytosine
Deoxyribose sugar
Ribose sugar
DNA TO PROTEIN
Protein is made of amino acid sequences
20 amino acids
How does DNA code for amino acid?
GENETIC CODE
Codon
Three
letter code
5’ to 3’ order
Start codon
Stop codon
AA are represented
by more than one
codon
61 codons that
specify AA
AMINO ACIDS
Abbreviated
Three
letters
RNA polymerase
TRANSCRIPTION
DNA to RNA
Occurs in nucleus
Three process
Initiation
DNA of gene
Promoter
DNA
Initiation
Terminator
DNA
Elongation
Elongation
Termination
Termination
Growing
RNA
Completed RNA
RNA
polymerase
INITIATION
RNA polymerase
binds to DNA
Binds at promoter
region
TATA
box
RNA polymerase
unwinds DNA
Transcription unit
Part
of gene that is
transcribed
INITIATION
Transcription factors bind
to specific regions of
promoter
Provide a substrate for
RNA polymerase to bind
beginning transcription
Forms transcription
initiation complex
ELONGATION
RNA molecule is built
RNA polymerase
Primer not needed
5’ to 3’
3’ to 5’ DNA is template
strand
Coding strand
Produces mRNA
DNA strand that is not
copied
Messenger RNA
DNA double helix
reforms
TERMINATION
RNA polymerase recognizes a termination
sequence – AAAAAAA
Nuclear proteins bind to string of UUUUUU on
RNA
mRNA molecule releases from template strand
POST-TRANSCRIPTIONAL MODIFICATIONS
Pre-mRNA undergoes
modifications before it
leaves the nucleus
Poly(A) tail
Poly-A polymerase
Protects from RNA
digesting enzymes in
cytosol
5’ cap
7 G’s
Initial attachment site for
mRNA’s to ribosomes
Removal of introns
SPLICING THE PRE-MRNA
DNA comprised of
Exons
– sequence of
DNA or RNA that
codes for a gene
Introns – non-coding
sequence of DNA or
RNA
Spliceosome
Enzyme
that removes
introns from mRNA
SPLICING PROCESS
Spliceosome contains a handful of small
ribonucleoproteins
snRNP’s
(snurps)
snRNP’s bind to specific regions on introns
ALTERNATIVE SPLICING
Increases number and variety of proteins
encoded by a single gene
~25,000 genes produce ~100,000 proteins
TRANSLATION
mRNA to protein
Ribosomes read
codons
tRNA assists
ribosome to
assemble amino
acids into
polypeptide chain
Takes place in
cytoplasm
TRNA
Contains
triplet
anticodon
amino acid attachment
site
Are there 61 tRNA’s to
read 61 codons?
TRNA: WOBBLE HYPOTHESIS
First two nucleotides of codon for a specific AA is always
precise
Flexibility with third nucleotide
Aminoacylation – process of adding an AA to a tRNA
Forming aminoacyl-tRNA molecule
Catalyzed by 20 different aminoacyl-tRNA synthetase enzymes
RIBOSOMES
Translate mRNA chains into amino acids
Made up of two different sized parts
Ribosomal
subunits (rRNA)
Ribosomes bring together mRNA with
aminoacyl-tRNAs
Three sites
A
site - aminoacyl
P site – peptidyl
E site - exit
Amino acid
TRANSLATION
PROCESS
Polypeptide
A
site
P site
Anticodon
mRNA
1
Three stages
Initiation
Elongation
Codon recognition
mRNA
movement
Termination
Stop
codon
New
peptide
bond
3
Translocation
2
Peptide bond
formation
INITIATION
Ribosomal subunits associate with mRNA
Met-tRNA (methionine)
Forms complex with ribosomal subunits
Complex binds to 5’cap and scans for start codon (AUG) –
known as scanning
Large ribosomal subunit binds to complete ribosome
Met-tRNA is in P-site
Reading frame
is established
to correctly
read codons
ELONGATION
Amino acids
are added to
grow a
polypeptide
chain
A, P, and E
sites operate
4 Steps
TERMINATION
A site arrives at a stop codon on mRNA
UAA, UAG, UGA
Protein release factor binds to A site releasing
polypeptide chain
Ribosomal subunits, tRNA release and detach from
mRNA
POLYSOME
b
a
Red object = ?
What molecules are present in this
photo?
PROKARYOTIC RNA TRANSCRIPTION/TRANSLATION
Throughout cell
Single type of RNA
polymerase transcribes
all types of genes
No introns
mRNA ready to be
translated into protein
mRNA is translated by
ribosomes in the cytosol
as it is being transcribed
REVIEW
What is a gene?
Where is it located?
What is the main
function of a gene?
Do we need our genes
“on” all the time?
How do we turn genes
“on” or “off”?
REGULATING GENE EXPRESSION
Proteins are not required by
all cells at all times
Regulated
Eukaryotes – 4 ways
Transcriptional (as mRNA is
being synthesized)
Post-transcriptional (as mRNA
is being processed)
Translational (as proteins are
made)
Post-translational (after
protein has been made)
Prokaryotes
lacOperon
trpOperon
TRANSCRIPTIONAL REGULATION
Most common
DNA wrapped around histones keep gene promoters
inactive
Activator molecule is used (2 ways)
Signals a protein remodelling complex which loosen the
histones exposing promoter
Signals an enzyme that adds an acetyl group to histones
exposing promoter region
TRANSCRIPTIONAL REGULATION
Methylation
Methyl groups are added to the cytosine bases in the
promoter of a gene (transcription initiation complex)
Inhibits transcription – silencing
Genes are placed “on hold” until they are needed
E.g. hemoglobin
AGOUTI MICE
POST TRANSCRIPTIONAL REGULATION
Pre-mRNA processing
Alternative splicing
Rate of mRNA degradation
Masking proteins – translation
does not occur
Embryonic
development
Hormones - directly or
indirectly affect rate
Casein – milk protein in
mammary gland
When casein is needed,
prolactin is produced
extending lifespan of casein
mRNA
TRANSLATIONAL REGULATION
Occurs during protein synthesis by a ribosome
Changes in length of poly(A) tail
Enzymes
add or delete adenines
Increases or decreases time required to translate
mRNA into protein
Environmental
cues
POST-TRANSLATIONAL REGULATION
Processing
Removes
sections of protein to make it active
Cell regulates this process (hormones)
Chemical modification
Chemical
groups are added or deleted
Puts the protein “on hold”
Degradation
Proteins
tagged with ubiquitin are degraded
Amino acids are recycled for protein synthesis
PROKARYOTIC REGULATION
lacOperon
Regulates the production of lactose metabolizing proteins
PROKARYOTIC REGULATION
trpOperon
Regulates the expression of tryptophan enzymes
CANCER
Lack regulatory mechanisms
Mutations in genetic code (mutagens)
Probability
increases over lifetime
Radiation, smoking, chemicals
Mutations are passed on to daughter cells
Can
lead to a mass of undifferentiated cells (tumor)
Benign and malignant
Oncogenes
Mutated
genes that once served to stimulate cell
growth
Cause undifferentiated cell division
CANCER
GENETIC MUTATIONS
Positive and negative
Natural
selection – evolution
Cancer –death
Small-Scale – single base pair
Point
mutations
Substitution,
insertion/deletion, inversion
Large-Scale – multiple base pairs
SMALL-SCALE MUTATIONS
Four groups
Missense,
Lactose,
SNPs
nonsense, silent, frameshift
sickle cell anemia
– single nucleotide polymorphisms
Caused
by point mutations
MISSENSE MUTATION
Change of a single base pair or group of base pairs
Results in the code for a different amino acid
Protein will have different sequence and structure
and may be non-functional or function differently
NONSENSE MUTATION
Change in single base pair or group of base
pairs
Results in premature stop codon
Protein will not be able to function
SILENT MUTATION
Change in one or more base pairs
Does not affect functioning of a gene
Mutated DNA sequence codes for same amino
acid
Protein is not altered
FRAMESHIFT MUTATION
One or more nucleotides are inserted/deleted from a
DNA sequence
Reading frame of codons shifts resulting in multiple
missense and/or nonsense effects
Any deletion or insertion of base pairs in multiples of
3 does not cause frameshift
LARGE-SCALE MUTATIONS
Multiple
nucleotides, entire
genes, whole
regions of
chromosomes
LARGE-SCALE MUTATIONS
Amplification – gene
duplication
Entire
genes are copied
to multiple regions of
chromosomes
LARGE-SCALE MUTATIONS
Large-scale deletions
Entire
coding regions of DNA are removed
Muscular
Dystrophy
LARGE-SCALE MUTATIONS
Chromosomal translocation
Entire
genes or groups of genes are moved from
one chromosome to another
Enhance, disrupt expression of gene
LARGE-SCALE MUTATIONS
Inversion
Portion
of a DNA molecule reverses its direction in the
genome
No direct result but reversal could occur in the middle
of a coding sequence compromising the gene
LARGE-SCALE MUTATIONS
Trinucleotide repeat expansion
Increases
number of repeats in genetic code
CAG CAG CAG CAG CAG CAG CAG CAG
Huntingtons
disease
CAUSES OF GENETIC MUTATIONS
Spontaneous mutations
Inaccurate
DNA replication
Induced mutations
Caused
by environmental agent – mutagen
Directly alter DNA – entering cell nucleus
Chemicals, radiation
CHEMICAL MUTAGENS
Modify individual nucleotides
Nucleotides
resemble other base pairs
Confuses replication machinery – inaccurate copying
Nitrous
acid
Mimicking DNA nucleotides
Ethidium
bromide – insert itself into DNA
RADIATION - LOW ENERGY
UV B rays
Non-homologous end joining
Bonds form between adjacent nucleotides along DNA
strand
Form kinks in backbone
Skin cancer
RADIATION – HIGH ENERGY
Ionizing radiation – x-ray, gamma rays
Strip molecules of electrons
Break bonds within DNA
Delete portions of chromosomes
Development of tumors
MUTATION IN PROKARYOTES
DNA is mostly coding sequences
Mutation is harmful – superbugs
GENOMES AND GENE ORGANIZATION
Human Body
22
autosomal chromosomes
1 pair of each sex chromosome (XX, YY)
GENOMES AND GENE ORGANIZATION
Components
VNTR’s–variable
number tandem repeats (microsatellites)
Sequences of long repeating base pairs
TAGTAGTAGTAGTAG
LINEs
– long interspersed nuclear elements
SINEs – short interspersed nuclear elements
Transposons – small sequences of DNA that move
about the genome and insert themselves into different
chromosomes
Pseudogene – code is similar to gene but is unable to
code for protein
VIRUSES
Not alive but can replicate themselves
Contain
DNA
or RNA
Capsid – protein coat
Envelope – cell membrane
VIRUS
4000 species of virus have been classified
REPLICATION
DNA
Transcription
and translation
RNA (retrovirus)
Uses
reverse transcriptase – enzyme
Uses cells parts to make a single strand of DNA and
then makes a complementary strand from that copy
Integrase – incorporates into our genetic code
VIRUS AS VECTORS
Transduction
Using
a virus vector to insert DNA into a cell or
bacterium