P-type
advertisement
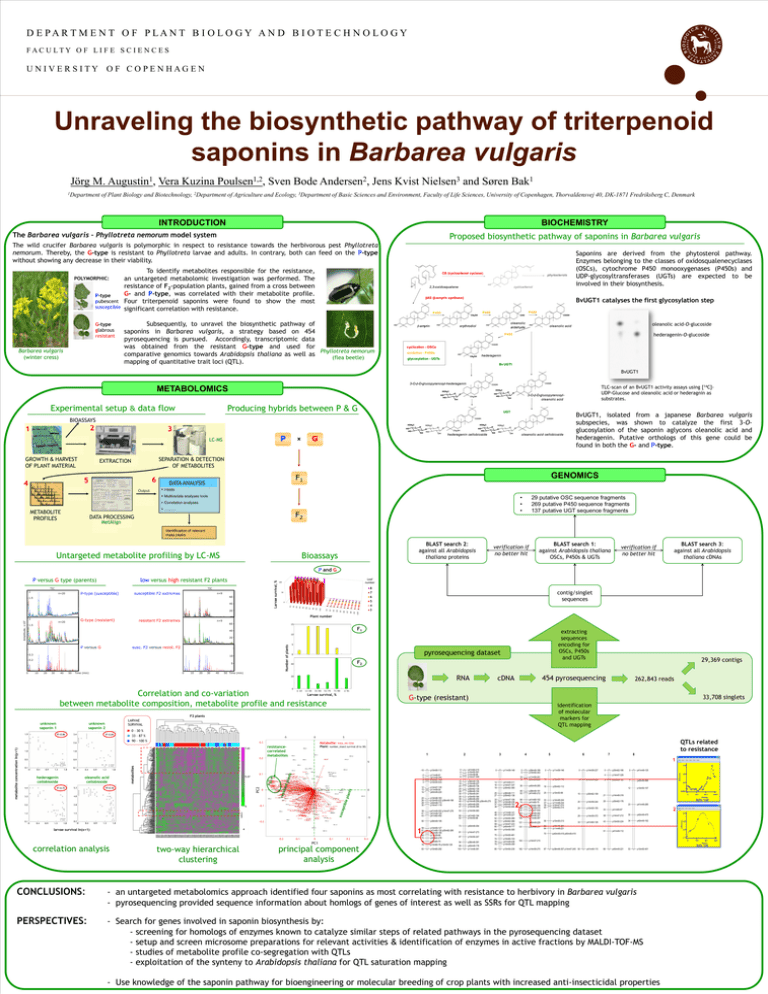
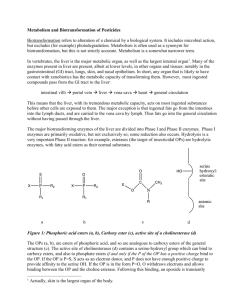
D E PAR T M E N T O F P LAN T B I O L O G Y AN D B I O T E C H N O L O G Y FACULTY OF LIFE S CIENC ES UNIVERSITY OF COPENHAGEN Unraveling the biosynthetic pathway of triterpenoid saponins in Barbarea vulgaris Jörg M. Augustin1, Vera Kuzina Poulsen1,2, Sven Bode Andersen2, Jens Kvist Nielsen3 and Søren Bak1 1Department of Plant Biology and Biotechnology, 2Department of Agriculture and Ecology, 3Department of Basic Sciences and Environment, Faculty of Life Sciences, University of Copenhagen, Thorvaldensvej 40, DK-1871 Fredriksberg C, Denmark INTRODUCTION BIOCHEMISTRY The Barbarea vulgaris – Phyllotreta nemorum model system Proposed biosynthetic pathway of saponins in Barbarea vulgaris The wild crucifer Barbarea vulgaris is polymorphic in respect to resistance towards the herbivorous pest Phyllotreta nemorum. Thereby, the G-type is resistant to Phyllotreta larvae and adults. In contrary, both can feed on the P-type without showing any decrease in their viability. POLYMORPHIC: P-type pubescent susceptible G-type glabrous resistant Barbarea vulgaris (winter cress) To identify metabolites responsible for the resistance, an untargeted metabolomic investigation was performed. The resistance of F2-population plants, gained from a cross between G- and P-type, was correlated with their metabolite profile. Four triterpenoid saponins were found to show the most significant correlation with resistance. CS (cycloartenol cyclase) phytosterols 2,3-oxidosqualene cycloartenol βAS (β-amyrin synthase) BvUGT1 catalyses the first glycosylation step P450 P450 P450 Subsequently, to unravel the biosynthetic pathway of saponins in Barbarea vulgaris, a strategy based on 454 pyrosequencing is pursued. Accordingly, transcriptomic data was obtained from the resistant G-type and used for Phyllotreta nemorum comparative genomics towards Arabidopsis thaliana as well as (flea beetle) mapping of quantitative trait loci (QTL). β-amyrin Saponins are derived from the phytosterol pathway. Enzymes belonging to the classes of oxidosqualenecyclases (OSCs), cytochrome P450 monooxygenases (P450s) and UDP-glycosyltransferases (UGTs) are expected to be involved in their biosynthesis. oleanolic aldehyde erythrodiol oleanolic acid-O-glucoside oleanolic acid hederagenin-O-glucoside P450 cyclisation - OSCs oxidation - P450s hederagenin glycosylation - UGTs Bv UGT1 BvUGT1 3-O-β-D-glucopyranosyl-hederagenin TLC-scan of an BvUGT1 activity assays using [14C]UDP-Glucose and oleanolic acid or hederagnin as substrates. METABOLOMICS 3-O-β-D-glucopyranosyloleanolic acid Experimental setup & data flow Producing hybrids between P & G UGT BIOASSAYS 2 1 3 P LC-MS GROWTH & HARVEST OF PLANT MATERIAL METABOLITE PROFILES G x oleanolic acid cellobioside SEPARATION & DETECTION OF METABOLITES EXTRACTION 6 5 4 hederagenin cellobioside BvUGT1, isolated from a japanese Barbarea vulgaris subspecies, was shown to catalyze the first 3-Oglucosylation of the saponin aglycons oleanolic acid and hederagenin. Putative orthologs of this gene could be found in both the G- and P-type. GENOMICS F1 DATA ANALYSIS • • • F2 DATA PROCESSING MetAlign Untargeted metabolite profiling by LC-MS BLAST search 2: against all Arabidopsis thaliana proteins Bioassays 29 putative OSC sequence fragments 269 putative P450 sequence fragments 137 putative UGT sequence fragments verification if no better hit BLAST search 1: against Arabidopsis thaliana OSCs, P450s & UGTs BLAST search 3: against all Arabidopsis thaliana cDNAs verification if no better hit P and G 2 Amplitude, ×107 resistant F2 extremes G9 contig/singlet sequences G10 G7 G8 G5 G6 G3 G4 G1 G2 Plant number G-type (resistant) n=20 P9 20 P10 0 P7 40 P8 60 P5 n=9 P6 susceptible F2 extremes P3 P-type (susceptible) n=20 1.5 8 7 6 5 4 3 50 P4 TIC P1 TIC 2 Leaf number 100 P2 low versus high resistant F2 plants Larvae survival, % P versus G type (parents) n=9 60 1.5 15 F1 40 extracting sequences encoding for OSCs, P450s and UGTs 10 20 susc. F2 versus resist. F2 0.3 Intens. x10 8 Number of plants P versus G 1.5 1.0 10 0.5 5 0.2 0.0 0 10 20 30 40 50 Time (min) 0 0 10 10 20 20 30 30 40 40 50 Time (min) 50 5 pyrosequencing dataset 0 F2 40 Time [min] RNA 20 0 Correlation and co-variation between metabolite composition, metabolite profile and resistance ≤ 10 11-30 31-50 51-70 71-90 454 pyrosequencing cDNA G-type (resistant) 33,708 singlets identification of molecular markers for QTL mapping F2 plants LARVAE SURVIVAL unknown saponin 2 0 – 30 % 33 – 87 % -5 90 – 100 % 0 0.3 7.65 0.2 5 QTLs related to resistance Metabolite: mass_ret.time Plant: number_insect survival (0 to 30) resistancecorrelated metabolites 1 2 3 4 5 6 7 8 1 5 oleanolic acid cellobioside 5.07 0 3 12 4 0 0 -0.1 1 2 3 -5 -0.2 larvae survival ln(x+1) -0.2 two-way hierarchical clustering p10m50-112 -3 0 2 3 p11m48-81 p41m49-350 p14m47-319 p10m50-433 12 13 14 p14m47-130 p11m59-54 p40m49-220 24 26 28 30 p14m48-163 p10m50-222 p38m49-196 p32m48-172 p14m47-119 39 41 43 p41m49-302 p14m47-276 p41m49-124 p41m60-492 50 p38m49-195 1 4 correlation analysis -10 0.1 PC2 hederagenin cellobioside metabolites metabolite concentration ln(y+1) 262,843 reads ≥ 91 Larvae survival, % unknown saponin 1 29,369 contigs -0.1 0 PC1 0.1 0.2 principal component analysis 0.3 59 61 64 66 70 74 78 p10m50-127 p41m60-130 p35m62-280 p32m48-185 p11m59-66 p35m62-278 p15m60-75 p15m60-76 p10m50-126 83 p15m60-292 0 4 7 8 p41m60-215 p10m50-301 p14m48-88 p14m47-309 15 19 20 25 26 30 35 38 40 45 47 p38m49-135 p14m48-227 p38m49-136 p35m62-139 p38m48-104 p16m49-341 p10m50-164 p38m48-200 p14m48-256 p38m49-275 p40m49-255 p14m48-394 53 p14m59-383 60 62 p41m60-194 p10m50-88 73 p40m49-354 81 p14m47-273 87 p10m50-247 94 p38m48-391 99 103 p40m49-178 p11m48-391 0 13 16 19 0 5 p38m49-189 p10m50-343 0 p11m59-146 18 19 p11m59-251 p40m49-266 10 p14m59-110 36 p40m49-265 18 p35m62-112 49 52 p38m49-191 p41m60-212 25 p10m50-98 68 74 83 86 91 102 105 108 p41m60-102 p41m60-70 p15m60-198 p11m59-215 p14m47-434 p14m59-340 p38m48-321 p11m48-307 121 p40m49-229 131 p40m49-231 p11m59-148 p41m49-311 p14m47-337 p35m62-365 25 27 p38m48-111 p35m62-138 33 34 37 38 p14m47-106 p14m48-238 p40m49-183 p10m50-468 46 48 52 55 59 60 64 p11m48-141 p41m49-227 p10m50-369 p35m62-369 p14m47-223 p40m49-117 p10m50-305 70 p10m50-461 74 78 79 p15m60-103 p41m49-436 p14m59-309 163 p14m47-215 85 p10m50-230 182 p14m59-270 2 33 37 39 40 p14m48-244 p11m48-234 p14m47-210 p35m62-214 0 6 p14m59-227 p14m47-296 15 p35m62-109 20 p14m59-244 24 p10m50-155 29 33 56 p10m50-213 62 64 p40m49-294 p11m48-221 70 p40m49-418 p40m49-419 87 p38m48-307 p14m47-245 50 0 p35m62-166 4 p14m47-326 8 p40m49-180 21 p14m48-318 26 p35m62-176 0 p41m49-125 7 p40m49-466 12 p10m50-107 23 p41m49-285 2 33 p41m60-87 p14m59-372 38 p14m47-212 30 p40m49-475 p41m60-126 43 p10m50-224 34 p40m49-162 51 p41m49-112 66 p40m49-221 53 p10m50-401 p41m49-115 CONCLUSIONS: - an untargeted metabolomics approach identified four saponins as most correlating with resistance to herbivory in Barbarea vulgaris - pyrosequencing provided sequence information about homlogs of genes of interest as well as SSRs for QTL mapping PERSPECTIVES: - Search for genes involved in saponin biosynthesis by: - screening for homologs of enzymes known to catalyze similar steps of related pathways in the pyrosequencing dataset - setup and screen microsome preparations for relevant activities & identification of enzymes in active fractions by MALDI-TOF-MS - studies of metabolite profile co-segregation with QTLs - exploitation of the synteny to Arabidopsis thaliana for QTL saturation mapping - Use knowledge of the saponin pathway for bioengineering or molecular breeding of crop plants with increased anti-insecticidal properties