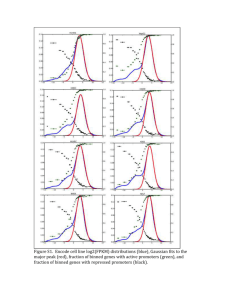
Final
advertisement

The development of a RNA-sequencing pipeline based on tuxedo tools Martijn Derks Masoed Ramuz Nick Alberts Rico Hagelaar Index • • • • • • Dataset Pipeline 1 (Tophat_cuff) Pipeline 2 (Cuff_diff) Pipeline 3 (Summary) Conclusions Future prospects Dataset • Arabidopsis thaliana (advanced) • Six conditions: • • • • • • Cold stress Drought stress Heat stress Highlight stress Salt stress Control Gan et al. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 477, P 419–423. Tophat_cuff Input data (FastQ) Configuration file Tophat (6x ) Bamfile Cufflinks Transcripts.gtf Analysis Total intron length Transcript length Basic for plot R Tophat_cuff results Cold Drought Heat highlight Salt WT mapped 11.01M 10.63M 11.24M 10.96M 7.41M 20.11M unmapped 23.90M 25.18M 21.82M 19.97M 24.30M 33.64M percentage 31.5 29.7 34.0 35.4 23.4 37.4 Tophat_cuff results Condition # genes FPKM > 1 Cold_stress 34029 20348 Drought_stress 35060 21044 Heat_stress 33615 19079 Highlight_stress 38480 22557 Salt_stress 33778 20111 Cuff_diff (1) (5x ) Control vs condition transcript.gtf Cuffmerge Merged.gtf Bamfile Cuffdiff DE-genes Functions + enrichment Cuff_diff (2) uniprot Get Functions David Enrichment (5x ) Cuff_diff results (Uniprot) • XLOC_005119 XLOC_005119 Hsp70b 1:5502205-5504535 WT_control heat_stress OK 1.88554 4668.1 11.2736 -4.26394 2.00852e-05 0.00596 • 568 yes Q9S9N1 Heat shock 70 kDa protein 5 (Heat shock protein 70-5) (AtHsp70-5) (Heat shock protein 70b) FUNCTION: In cooperation with other chaperones, • Hsp70s stabilize preexistent proteins against aggregation and mediate the folding of newly translated polypeptides in the cytosol as well as within organelles. These • chaperones participate in all these processes through their ability to recognize nonnative conformations of other proteins. They bind extended peptide segments with a • net hydrophobic character exposed by polypeptides during translation and membrane translocation, or following stress-induced damage (By similarity). Cytopla • sm. ATP binding; cell wall; chloroplast; plasma membrane; response to heat; response to virus GO:0005524; GO:0005618; GO:0009507; GO:0005886; GO:0009408; GO: • 0009615 Cuff_diff results DE genes/overlap Cuff_diff results (David) Drought Cold Heat Salt Highlight Summary Summary ID Cold Drought Heat Highlight Salt WT AT1G01010 10.5501 12.0209 6.80685 6.44518 Tophat count AT_codes Csv maker 0 10.7992 GC genes vs FPKM AT1G01030 2.51058 2.60705 0.582286 3.71439 1.37225 2.46655 AT1G01046 0 0 0 6.40264 4.73081 0 AT1G01050 52.7297 75.5912 0 CV46.9862 0 46.5351 Expr. intron AT1G01070 13.6023 15.9691 0 7.52686 19.3891 23.0487 AT1G01073 0 0 0 0 0 0 AT1G01090 80.2276 70.2176 58.4497 67.0227 102.39 Conservation Overlap 80.5032 matrix Clustering R AT1G01110 0.966456 1.307 0.564864 1.26781 1.88932 2.65862 CV= STDEV/ Average HC sample Clustering HC gene Clustering 0.15 Heatmap Clustering HC clusters (9) PAM clusters (10) Transcription factors Abscisic acid biosynthesis (stress conditions) 1 2 1. Cold, salinity and drought stresses: An overview Shilpi Mahajan Narendra Tuteja 2. Cold stress regulation of gene expression in plants Viswanathan Chinnusamy et al. Conserved genes in Arabidopsis • Abiotic stress genes which also occur in Arabidopsis were retrieved from Oryza sativa (Rabbani et al). • These genes were compared with the DE stress genes found in the results. • Three genes were found in the salt, cold and drought conditions. • Rabbani, M.A. Maruyama, K. Abe, H. Khan, M. A. Katsura, K. Ito, Yoshiwara, K. Seki, M. Shinozaki, K. Yamaguchi-Shinozaki, K. 2003. Monitoring Expression Profiles of Rice Genes under Cold, Drought, and High-Salinity Stresses and Abscisic Acid Application Using cDNA Microarray and RNA Gel-Blot Analyses. Plant Physiology vol. 133. No 4. Pp 1755-1767 Literature overlap • Results of the GO enrichment are backed up by the literature, with the exception of high light stress • The crosstalk between drought, cold and salt stress was confirmed by the literature with a greater emphasis on drought and salt stress. Seki, M. Narusaka, M. Ishida, J. Nanjo, T. Fujita, M. Oono, Y. Kamiya, A. Nakajima, M. Enju, A. Sakurai, T. Satou, M. Akiyama, K. Taji, T. Yamaguchi-Shinozaki, K. Carninci, P. Kawai, J. Hayashizaki, Y. Shinozaki, K. 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. V 31. I 3. pp 279-292. Baniwal, K. S. Bharti, K. Yu Chan, K. Fauth, M. Ganguli, A. Kotak, S. Mishra, S. K. Nover, L. Port, M. Scharf, K. Tripp, J. Weber, C. Zielinski, D. Koskull-Doring, P. 2004. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. V 29. I 4. pp 471-487. Bartels, D. Nelson, D. 1994. Approaches to improve stress tolerance using molecular genetics. Plant, Cell and Environment. V 17. pp 659-667. Wang, W. Vinocur, B. Shoseyov, O. Altman, A. 2004. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. V 9. I 5. pp. 244-252. Conclusions • Working pipeline for (Paired + Unpaired) RNAseq analysis • DE genes + Gene Enrichment detection • Cluster analysis CV genes • Differential expressed genes identified (stress conditions vs. WT) • Correlation Transcript length with FPKM • Not found in Intron/GC percentage • Clusters of Co-expressed genes • Assumption of co-regulated genes Future perspectives • Use different IDs (TAIR IDs are not suitable) • Transcription factors to cluster genes (similar regulatory elements? ) • Conservation other plant species (synteny) • Validation different dataset (organisms, paired end) Questions