Comparative Genomics(ppt)
advertisement
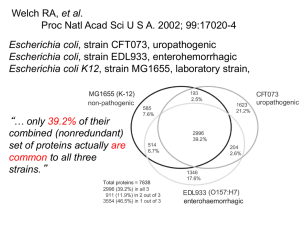
Comparative Genomics Todd Castoe Biochemistry and Molecular Genetics The First Genomes Figure 18.6 Genomes 3 (© Garland Science 2007) Tree of life from David Hillis’ lab (based on ~3000 rRNAs) animals plants you are here protists fungi http://www.zo.utexas.edu/faculty/antisense/Download.html bacteria archaea Tree of life from David Hillis’ lab (based on ~3000 rRNAs) you are here http://www.zo.utexas.edu/faculty/antisense/Download.html Hedges, Nat Rev Genet 2003 An argument for model species and the need for comparative genomics Most human proteins are ancient >90% Timescale of eukaryote evolution ~75% HUMAN PROTEINS… ~50% ~30% Divergences within 749 gene families in the Human Genome Gu X. et al. Nature Genetics (2002) 31 205-209 Genomes have been recycling for Billions of years What is comparative genomics There are many ways that genomes can be compared • Whole genome – Genome size – Genome alignments – Synteny (gene order conservation) – Gene number – Anomalous regions • Gene-centric – Gene families and unique genes – Gene clustering by function • Gene sequence variations – Codon usage, SNPs, inDels, pseudogenes 11 Why Comparative Genomics? 1. Conservation over long evolutionary distances suggests functional constraints 2. Lack of conservation over short distances may be indicative of adaptive evolution 3. Helps us identify both coding and non-coding genes and regulatory elements 4. Characterizing the differences between organisms reveals mechanisms of change 5. Allows us to achieve a greater understanding of vertebrate evolution 6. Leveraging knowledge between species for annotation and inference of function 7. Tells us what is common and what is unique between different species at the genome level 8. The function of human genes and other regions may be revealed by studying their counterparts in simpler model organisms 12 Comparing Genome Size The ‘C-value paradox’ Genome size does NOT correlate with organismal complexity 13 Why Are Some Genomes So Large? • There is no clear correlation between genome size and genetic complexity. • C-value – The total amount of DNA in the genome (per haploid set of chromosomes) • C-value paradox – The lack of relationship between the DNA content (C-value) of an organism and its coding potential. Haploid Genome Size (log scale) Contrasted Genome Landscapes Transposable Element The amount of TE correlate positively with genome size Mb Genomic DNA 3000 2500 TE DNA 2000 Protein-coding DNA 1500 1000 500 0 Feschotte & Pritham 2006 17 Transposable Elements… • Variation in gene numbers cannot explain variation in genome size among eukaryotes • Most of variation in genome size is due to variation in the amount of repetitive DNA (mostly derived from TEs) • TEs accumulate in intergenic and intronic regions •CONCLUSIONS… •TEs have played an important role in genome evolution and diversification •Facilitate expansion and contraction of genomes AND gene families 18 Coarse Comparisons of Genomes 19 Fugu Genome Science 2002 365 Mb (1/10 the human) Tiny vertebrate genome Humans and Fish shared common ancestor 450Mya! 20 Among the Smallest Vertebrate Genome • Genome is < 1/6 repetitive DNA – Vs. ~50% in us • ¾ of human proteins have a strong match to Fugu (pretty good for 450My) • ¼ of human proteins had highly diverged from, or had no pufferfish homologs 21 Shadows of the Ancient Vertebrate Genome… • Conserved linkages between Fugu and human – Preservation of chromosomal chunks from the common vertebrate ancestor (synteny) • BUT, lots of cut/copy-paste…. And some general scrambling of gene order 22 Shadows of the Ancient Vertebrate Genome… • Conserved linkages between Fugu and human – Preservation of chromosomal chunks from the common vertebrate ancestor • BUT, lots of cut/copy-paste…. And some general scrambling of gene order What a little genome… …with little introns • The Fugu genome is compact partly because introns are shorter compared with the human genome • The Fugu mode of intron size is 79 bp – 75% of introns 425 bp in length • The human mode is 87 bp – 75% of introns 2609 bp • Fugu: 500 introns > 10Kb --- Human: 12,000 > 10Kb • The total numbers of introns are roughly the same – 161,536 introns in Fugu – 152,490 introns in human What a little genome… …with little introns GC Content Differences Probably related to the relative complexity of the chromatin structure in humans versus the Fugu. Fugu-Human Synteny http://blast.fugu-sg.org/fugu-synteny/viewer_newServer.php I think their maps, however, are confusing and not that informative, -scaffolds were not physically mapped to chromosomes… Let’s look instead at the other pufferfish, Tetraodon, that was sequenced the following year.. -physical mapping to chromosomes was complete Tetraodon-Human Synteny 28 Comparative Genomics – Synetny Human Chrom.1 vs. Chimp 29 Comparative Genomics – Synetny Human Chrom.1 vs. Mouse 30 Comparative Genomics – Synetny Human Chrom.1 vs. Cow 31 Comparative Genomics – Synetny Human Chrom.1 vs. Opossum 32 Comparative Genomics – Synetny Human Chrom.1 vs. Platypus 33 Comparative Genomics – Synetny Human Chrom.1 vs. Chicken 34 Synteny • Large blocks of synteny exist even at great phylogenetic distance • Also substantial scrambling, even at short distance… 35 Whole Genome Alignments • Functional sequences often evolve more slowly than non-functional sequences, therefore sequences that remain conserved may perform a biological function. • Comparing genomic sequences from species at different evolutionary distances allows us to identify: – Coding genes – Non-coding genes – Non-coding regulatory sequences 36 The Rate of Evolution Depends on Constraints Human vs. Rodent Comparison Highest substitution rates: pseudogenes introns 3’ flanking (not transcribed to mature mRNA) 4-fold degenerate sites Intermediate substitution rates: 5’ flanking (contains promoter) 3’, 5’ untranslated (transcribed to mRNA) 2-fold degenerate sites Lowest substitution rates: Nondegenerate sites Selection of Species for DNA comparisons Human vs.. Chimpanzee Mouse Opossum Pufferfish Size (Gbp) 3.0 2.5 4.2 0.4 Time since divergence ~5 MYA ~ 65 MYA ~150 MYA ~450 MYA Sequence conservation (in coding regions) >99% ~80% ~70-75% ~65% Aids identification of… Recently changed sequences and genomic rearrangements Both Both Primarily coding and coding and coding non-coding non-coding sequences sequences sequences 38 Comparative Analyses of Sequence Conservation Hypothesis: areas with high sequence similarity are likely to contain functionally important elements: protein-coding exons transcription factor binding sites These two are conceptually the same… Phylogenetic Shadowing (fine scale) Identifying regions that do not accumulate change Phylogenetic Footprinting (large scale) Identifying which regions stay somewhat conserved (identifiable) across larger evolutionary distances 39 UCSC Genome Browser 40 In these comparative genomic charts, it is easy to see why meaningful comparisons between humans and other primates have been difficult. The pink areas represent regions of high conservation between the two species being compared, (meaning the sequences are the same in both), the blue areas represent the positions of protein-coding regions and the purple areas represent the non-protein coding parts of a gene. 41 Phylogenetic shadowing analyses sequence variation in a multiple alignment to identify regions that accumulate variation at a slower rate. Each position of an alignment is fitted to a phylogenetic model to calculate the likelihood that the position is evolving at a fast or a slow rate (a). Generally, positions with several sequence differences across species are more likely to be evolving at a fast rate, and in turn identify the least variable regions (b). The slowly evolving regions often correspond to functional sequences. 42 Phylogenetic Footprinting (VISTA) 43 Identification of Conserved Regulatory Elements 44 Comparative analysis of multi-species sequences from targeted genomic regions Nature, 2003 45 CFTR Locus Encodes the protein: Cystic Fibrosis Transmembrane Conductance Regulator – An ion channel across the cell membrane – The transport of chloride through CFTR helps control the movement of water in tissues and maintain the fluidity of mucus and other secretions – Normal functioning ensures that organs such as the lungs and pancreas function properly – Most CF patients show a deletion that either leads to an amino acid substitution, or a deletion of part of an exon of CFTR Comparative Genomics of the CFTR Locus • CFTR = 1.8 Mb of human Ch7, Sequenced for 12 ssp. • How does a single locus change over evolutionary time? • How much does it change? • What types of changes are more/less common? • Do some lineages have more of certain changes than others? • How much comparative genomic data do we need??? 47 Sequence Conservation 48 Looking backward from the human genome How much is still there after 450my (Fugu) 49 Differences in exon length Data like this sure makes you wonder about mouse models of human disease, eh? Differences in exon lengths: + = insertion -= deletion e = extension due to alteration of splice site or stop codon s = early stop codon Transposable Elements Gone Wild! High Turnover in TEs despite gene conservation 51 Nucleotide Changes Big insertions/deletions More common Than nucleotide changes! In primates, large indels are the principal mechanism accounting for the observed sequence differences 52 Using evolutionary conservation to ID functionally important conserved human genome segments How many comparative genomes do we need – can’t we just use the mouse? (Lots, and NO)… Using all 12 species, they found 561 Multi-Species Conserved Sequences (MCSs) So, how many could we find using just the Mouse genome (rather than all 12) False Pos. True Pos. False Neg. Less than half even with high false positives…!!! 53 Multi-Species Conserved Sequences 950 of the 1,194 MCSs are neither exonic nor lie less than 1-kb upstream of transcribed sequence. Meaning they are otherwise hard to predict (= Evolutionary Distance) Strong argument for comparative genomics: Need many species, and distant species – like cat, dog, fish - to ID conserved possibly-functional regions in humans! 54 Take Home Messages… • Identification of conserved non-coding segments beyond those previously identified experimentally, and evidence we can find more with even more genomes!!! • These were not detectable by pair-wise sequence comparisons alone – Underscores importance of comparative genomics • Need many diverse species to figure out these questions! • Analysis of TE insertions highlights variation in genome dynamics among species – The rate of TE evolutionary dynamics in vertebrates is amazing, and hugely important for the structure and evolution of the genome • Importance of large insertion-deletion (not necessarily nucleotide changes) between closely related species, including humans and primates 55 ENCODE Project • Cross-reference existing with new data on human genome function • Identify the functional relevance of as many bases of human genome as possible. 56 ENCODE Project Findings (2007) • A total of 5% of the bases in the genome can be confidently identified as being under evolutionary constraint in mammals • For ~60% of these conserved bases, evidence of function based on experimental assays • However, not all bases within known functional regions are evolutionarily conserved • Much of the variation, while functional, appears to be evolving under little selective constraint! – While functional, must not be important enough for “fitness” to be highly conserved…. 57 Evolutionarily Conserved Regions 58 Comparative Genomics Where do babies come from? (ask your parents) Where do genes come from? Evolution of Gene Families in Vertebrates 59 Gene Duplication Orthologous genes: in different organisms, diverged from common ancestral gene by speciation A1 – A2 or B1 – B2 Paralogous genes: originated from common ancestral gene via gene duplication A1 – B1 or A1 – B2, etc… Homologs: genes that have the same ancestor Orthologues and Paralogues The Fate of Gene Duplicates Functional Conservation – both copies can retain original function Gene Loss – one (or both) copies can be lost either by complete deletion or by mutation leading to a pseudogene (non-functional copy) Neofunctionalization – e.g., one copy may take on a new function while the other copy retains the original function Subfunctionalization - each copy becomes specialized for a subset of ancestral gene’s roles (Hox genes seem to be an example) Humans genome duplication Van de Peer et al. Nature Reviews Genetics (2009) Gene Duplication Most gene families are small; exceptions often have an adaptive basis: immunoglobulin genes (1000 copies in humans), olfactory receptor genes (100’s of copies in mammals) Rho GTPases – Molecular Switches Control cytoskeletal architecture, survival, adhesion, proliferation, motility, etc. 65 Gene Gain and Loss…. In 550MY Sea urchin is estimated to have 23,300 genes with representatives of nearly all vertebrate gene families •Gene families are not as large as in vertebrates •Some genes thought to be vertebrate-specific were found in the sea urchin •Others were identified in sea urchin but not the chordate lineage, which suggests loss in the vertebrates. •The sea urchin has orthologs of genes associated with •Vision •Hearing •Balance •chemosensation in vertebrates •raw material for current vertebrate complex sensory gene programs).. 66 Expansion of urchin-specific Rho GTPases 67 Gain and loss of genes in gene families GAIN LOSS Human genome has 689 genes not present in the chimp and the chimp has 729 genes not present in humans. Demuth et al., 2006, PLoS 1 Despite expansion-contraction of gene families, there is little novel gain or complete loss Opossum genome… 180MY of change • The opossum genome contains ~18,000–20,000 protein-coding genes, the vast majority have eutherian orthologues. • Lineage-specific genes largely originate from expansion and rapid turnover in gene families involved in immunity, sensory perception and detoxification. • Only eight currently have strong evidence of representing functional genes without homologues in humans! 69 Conclusions • Studying biology and medicine means studying recycled genomic material • Studying evolution informs genomics – Studying genomics informs evolution • Knowing how genomes evolve can directly inform on how they function • More genomes = more data points for studying how they change through evolution, thus how they function 70