Screen - ChemAxon
advertisement
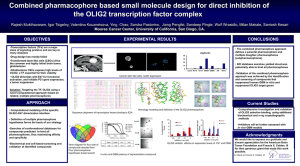
Screen Ligand based virtual screening presented by … maintained by Miklós Vargyas Last update: 13 April 2010 Screen Virtual screening by topological descriptors Screen Description of the product Screen performs high throughput virtual screening of compound libraries using similarity comparisons by various molecular descriptors. Availabilty • • • • • • • JChemBase JChem Oracle cartridge Instant Jchem Server version standalone command line application programs KNIME PipelinePilot Key features Various 2D descriptors • • • • • • ChemAxon chemical fingerprint (CCFP) PipelinePilot ECFP/FCFP ChemAxon pharmacophore fingerprint (CPFP) BCUT Scalars (logP, logD, Szeged index …) custom descriptors, in-house fingerprints Optimized similarity measures • • • Improves similarity prediction depends on set of known actives high enrichment ratios in virtual screening Multiple queries • • 3 types of hypotheses combined hit lists Benefits Versatile • Use various descriptors in your well established model • Access your trusted in-house fingerprint in IJC, JCB, JCART • Easy integration in corporate discovery pipelines • Search chemical files directly no need to import structures in database • New descriptors are pluggable in deployed systems Optimal • Consistent similarity scores • Smaller hit set • More focused library Benefits More consistent similarity scores 0.57 0.47 0.55 optimized Tanimoto 0.20 regular Tanimoto 0.28 0.06 Benefits High enrichment ratio • Fewer false hits • Known actives are true positive hits (ACE inhibitors) Number of Hits 10000 1000 100 10 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Number of Active Hits Tanimto Euclidean Optimized Ideal Results NPY-5 (pharmacophore similarity) Number of Hits 10000 1000 100 10 1 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49 Number of Spikes retrieved Tanimoto Euclidean Optimized Ideal Results β2-adrenoceptor (pharmacophore similarity) Number of Hits 10000 1000 100 10 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Number of Active Hits Tanimto Euclidean Optimized Ideal Case study at Axovan • GPCR activity prediction • distinguishing between GPCR subclasses GPCR-Tailored Pharmacophore Pattern Recognition of Small Molecular Ligands Modest von Korff and Matthias Steger, JCICS 2004, 44 Screen roadmap • New molecular descriptors – ECFP/FCFP (in 5.4) – Shape descriptors (in 5.4) • Hidden use of the optimiser – No-pain black-box approach – Simultaneous multi-descriptor search • Enhanced IJC integration – Easy descriptor configuration and generation – Similarity search type instead of descriptors, metrics and other unfriendly concepts Screen roadmap • GUI – New web interface (HTML/AJAX) – Desktop application for descriptor generation • 3D shape similarity – fast pre-filtering by 3D fingerprint – Alignment based volumetric Tanimoto calculation – scaffold hopping by maximizing topological dissimilarity and spatial similarity Supplementary slides A typical approach 0101010100010100010100100000000000010010000010010100100100010000 query fingerprint query B( x & y) B( x) B( y ) B( x & y ) metric 0000000100001101000000101010000000000110000010000100001000001000 0100010110010010010110011010011100111101000000110000000110001000 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0100011100011101000100001011101100110110010010001101001100001000 0101110100110101010111111000010000011111100010000100001000101000 0100010100111101010000100010000000010010000010100100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010011000000000000000000010100000010000000000000000000 0100010100010100000000000000101000010010000000000100000000000000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 0000000100000000010000100000000000001010100000000100000100100000 0100010100010100000000100000000000010000000000000100001000011000 0001000100001100010010100000010100101011100010000100001000101000 0100011100010100010000100001001110010010000010001100000000101000 0101010100010100010100100000000000010010000010010100100100010000 targets target fingerprints hits ChemAxon’s approach 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0101110100110101010111111000010000011111100010000100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010100000000000000101000010010000000000100000000000000 0101110100110101010111111000010000011111100010000100001000101000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 hypothesis fingerprint 0000000100000000010000100000000000001010100000000100000100100000 queries s min(x , y ) x s min(x , y ) 1 y s min(x , y ) s min(x , y ) i i i i i i i i i i i i i i i i i i i i optimized metric optimization 0000000100001101000000101010000000000110000010000100001000001000 0100010110010010010110011010011100111101000000110000000110001000 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0100011100011101000100001011101100110110010010001101001100001000 0101110100110101010111111000010000011111100010000100001000101000 0100010100111101010000100010000000010010000010100100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010011000000000000000000010100000010000000000000000000 0100010100010100000000000000101000010010000000000100000000000000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 0000000100000000010000100000000000001010100000000100000100100000 0100010100010100000000100000000000010000000000000100001000011000 targets 0001000100001100010010100000010100101011100010000100001000101000 0100011100010100010000100001001110010010000010001100000000101000 0101010100010100010100100000000000010010000010010100100100010000 target fingerprints hits Performance Chemical fingerprint generation: 500/s Pharmacophore fingerprint generation • calculated: 80/s • rule-based: 200/s Screening: 12000/s Optimization: 10s/metric Hardware/software environment: • P4 3GHz, 1GB RAM • Red Hat Linux 9 • Java 1.4.2 Implementations Use of various fingerprints and metrics in JSP http://www.chemaxon.com/jchem/examples/jsp1_x/index.jsp UGM presentation by Aureus Pharma Improved Virtual Screening Strategies and Enrichment of Focused Libraries in Active Compounds Using TargetOriented Databases http://www.chemaxon.com/forum/viewpost2307.html Molecular similarity Chemical, pharmacological or biological properties of two compounds match. The more the common features, the higher the similarity between two molecules. Chemical Pharmacophore Similarity measures Quantitative assessment of similarity of structures • need a numerically tractable form • molecular descriptors, fingerprints, structural keys Sequences/vectors of bits, or numeric values that can be compared by distance functions, similarity metrics. E ( x, y ) n x i 1 i yi 2 T ( x, y ) B( x & y) B( x) B( y ) B( x & y ) Standard metrics DTanimoto x y min(x , y ) ( x, y) 1 max(x , y ) x y min(x , y ) i i i i i DEuclidean( x, y ) x i i i i i yi 2 i DTanimoto ( , ) = 0.68 ( , ) = 21.93 DEuclidean i i i i i i i Topological chemical fingerprint hashed binary fingerprint • encodes topological properties of the chemical graph: connectivity, edge label (bond type), node label (atom type) • allows the comparison of two molecules with respect to their chemical structure Construction 1. 2. 3. find all 0, 1, …, n step walks in the chemical graph generate a bit array for each walks with given number of bits set merge the bit arrays with logical OR operation Construction of chemical fingerprint H H H C C O H H length H walk bit array 0 C 1010000000 1 C–H 0001010000 1 C–C 0001000100 2 C–C–H 0001000010 2 C–C–O 0100010000 3 C–C–O–H 0000011000 ALL 1111011110 Chemical similarity 0100010100010100010000000001101010011010100000010100000000100000 0100010100010100010000000001101010011010100000000100000000100000 Topological pharmacophore fingreprint • encodes pharmacophore properties of molecules as frequency counts of pharmacophore point pairs at given topological distance • allows the comparison of two molecules with respect to their pharmacophore Construction 1. 2. 3. 4. perceive pharmacophoric features map pharmacophore point type to atoms calculate length of shortest path between each pair of atoms assign a histogram to every pharmacophore point pairs and count the frequency of the pair with respect to its distance Pharmacophore perception Rule based approach Rule 1: The pharmacophore type of an atom is an acceptor, if • it is a nitrogen, oxygen or sulfur, and • it is not an amide nitrogen or sulfur, and • it is not an aniline nitrogen, and • it is not a sulfonyl sulfur, and • it is not a nitro group nitrogen. Exceptions to simple rules sp2 atom n-cyano-methil piperidine exception extra rules large number of rules maintenance, performance Effect of pH pH = 7 pH = 1 pH pH specific rules large number of rules maintenance, performance Pharmacophore perception Calculation based approach Step 1: estimation of pKa allows the determination of the protonation state for ionizable groups at the given pH Step 2: partial charge calculation Pharmacophore perception Calculation based approach Step 3: hydrogen bond donor/acceptor recognition Step 4: aromatic perception Step 5: pharmacophore property assignment acceptor negatively charged acceptor acceptor and donor hydrophobic none Pharmacophore fingerprint 12 11 10 9 8 7 6 5 4 3 2 1 0 A A A A A A D D D D D D D D D D D D H H H H H H H H H H H H H H H H H H A A A A A A A A A A A A D D D D D D A A A A A A D D D D D D H H H H H H 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 12 11 10 9 8 7 6 5 4 3 2 1 0 A A A A A A D D D D D D D D D D D D H H H H H H H H H H H H H H H H H H A A A A A A A A A A A A D D D D D D A A A A A A D D D D D D H H H H H H 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 1 2 3 4 5 6 Pharmacophore type coloring: acceptor, donor, hydrophobic, none. Fuzzy smoothing 5 2 2 1 1 2 3 0 0 AA1 AA2 AA3 AA4 AA5 AA6 AA1 DE=1.41 AA2 AA3 AA4 AA5 AA6 AA5 AA6 DE=0.45 2 4 5 1 2 0 1 0 AA1 AA2 AA3 AA4 AA5 AA6 AA1 AA2 AA3 AA4 Virtual screening using fingerprints 0101010100010100010100100000000000010010000010010100100100010000 query fingerprint query metric 0000000100001101000000101010000000000110000010000100001000001000 0100010110010010010110011010011100111101000000110000000110001000 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0100011100011101000100001011101100110110010010001101001100001000 0101110100110101010111111000010000011111100010000100001000101000 0100010100111101010000100010000000010010000010100100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010011000000000000000000010100000010000000000000000000 0100010100010100000000000000101000010010000000000100000000000000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 0000000100000000010000100000000000001010100000000100000100100000 0100010100010100000000100000000000010000000000000100001000011000 0001000100001100010010100000010100101011100010000100001000101000 0100011100010100010000100001001110010010000010001100000000101000 0101010100010100010100100000000000010010000010010100100100010000 targets target fingerprints hits Multiple query structures 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0101110100110101010111111000010000011111100010000100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010100000000000000101000010010000000000100000000000000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 0000000100000000010000100000000000001010100000000100000100100000 queries 0101110100110101010111111000010000011111100010000100001000101000 hypothesis fingerprint metric 0000000100001101000000101010000000000110000010000100001000001000 0100010110010010010110011010011100111101000000110000000110001000 0100010100011101010000110000101000010011000010100000000100100000 0001101110011101111110100000100010000110110110000000100110100000 0100010100110100010000000010000000010010000000100100001000101000 0100011100011101000100001011101100110110010010001101001100001000 0101110100110101010111111000010000011111100010000100001000101000 0100010100111101010000100010000000010010000010100100001000101000 0001000100010100010100100000000000001010000010000100000100000000 0100010100010011000000000000000000010100000010000000000000000000 0100010100010100000000000000101000010010000000000100000000000000 0101010101111100111110100000000000011010100011100100001100101000 0100010100011000010000011000000000010001000000110000000001100000 0000000100000000010000100000000000001010100000000100000100100000 0100010100010100000000100000000000010000000000000100001000011000 0001000100001100010010100000010100101011100010000100001000101000 0100011100010100010000100001001110010010000010001100000000101000 0101010100010100010100100000000000010010000010010100100100010000 targets target fingerprints hits Hypothesis fingerprints Advantages • allows faster operation • compiles features common to each individual actives • reduces noise Hypothesis types Active 1 0 2 7 1 0 1 6 4 0 0 9 0 Active 2 1 6 0 4 3 3 1 2 2 0 5 1 Active 3 2 4 4 1 0 2 5 3 4 3 4 5 Minimum 0 2 0 1 0 1 1 2 0 0 4 0 Average 1 4 3.67 2 1 2 4 3 2 1.33 6 2 Median 1.5 4 5.5 1 0 2 5 3 3 0 5 3 Hypothesis fingerprints Advantages Disadvantages Minimum • strict conditions for hits if actives are fairly similar • false results with asymmetric metrics • misses common features of highly diverse sets • very sensitive to one missing feature Average • captures common features of more diverse active sets • less selective if actives are very similar Median • captures common features of more diverse active sets • specific treatment of the absence of a feature • less sensitive to outliers • less selective if actives are very similar The need for optimization Too many hits The need for optimization Inconsistent dissimilarity values 0.57 0.47 0.55 Parametrized metrics scaled ,asymmetric Tanimoto D s min(x , y ) ( x, y) 1 x s min(x , y ) 1 y s min(x , y ) s min(x , y ) i i i i i i i i i i i i i i i i i i 0,1 asymmetry factor si N scaling factor weighted, asymmetric DEuclidean ( x, y) wi xi yi 2 xi yi 0,1 asymmetry factor wi 0,1 weights wi 1 xi yi 2 xi yi i i Optimization of metrics Step 1 optimize parameters for maximum enrichment Step 2 validate metrics over an independent test set training set training set query set selected targets known actives test set test set Optimization of metrics Step 1 optimize parameters for maximum enrichment query set 1111100010000100001000101000 query fingerprint Target hits parametrized metric training set Active hits Optimization of metrics v1 v2 v3 vi vn potential variable value temporarily fixed value final value running variable value Optimization of metrics Step 2 validate metrics over an independent test set query set Target hits 1111100010000100001000101000 query fingerprint optimized metric test set Active hits Results of Optimization 1. Similar structures get closer 0.57 0.47 0.55 0.20 0.28 0.06 Results of Optimization 2. Hit set size reduced Active set: 18 mGlu-R1 antagonists Target set: 10000 randomly selected drug-like structures Euclidean Tanimoto Metric Basic Scaled Asymmetric Scaled Asymmetric Basic Normalized Asymmetric Normalized Weighted Normalized Weighted Asymmetric Normalized Enrichment Test Hits Target Hits 70.47 5.43 172.00 7.63 6.00 1101.71 99.36 5.29 106.00 11.94 5.86 731.14 5.59 5.43 1465.57 11.33 5.14 791.29 18.58 4.71 368.71 296.30 4.14 27.57 281.30 3.43 17.00 Results of Optimization 3. Higher enrichment Active set 5-HT3 ACE Angiotensin Beta2 D2 Delta FTP mGluR1 NPY-5 Thrombin size Euclidean Optimized Improvement 12 12.55 239.24 49.26 89 1.42 6.50 4.64 10 27.81 85.45 11.15 50 1.52 24.70 17.42 13 27.64 123.25 11.19 20 11.66 243.57 69.11 35 46.88 71.54 5.35 18 5.59 296.30 70.93 139 3.05 12.75 3.25 8 2.56 7.68 2.62 Results of Optimization 4. Top ranked structures are spikes • offers a more intuitive way to evaluate the efficiency of screening • based on sorting random set hits and known actives on dissimilarity values and counting the number of random set hits preceding each active in the sorted list number of virtual hits 0.014 0.015 0.017 0.020 0.022 0.023 0.027 0.041 0.043 number of spikes retrieved Results ACE (pharmacophore similarity) 10000 Number of hits 1000 Euclidean 100 Optimized Euclidean 10 1 1 2 3 4 5 6 7 8 9 10 11 Number of spikes retrieved 12 13 14 15 16 Results NPY-5 (pharmacophore similarity) Number of Hits 10000 1000 100 10 1 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49 Number of Spikes retrieved Tanimoto Euclidean Optimized Ideal Results β2-adrenoceptor (pharmacophore similarity) Number of Hits 10000 1000 100 10 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Number of Active Hits Tanimto Euclidean Optimized Ideal 3D flexible search Expected top performance 200 structures/s