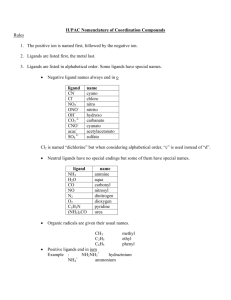
Composti di coordinazione: nomenclatura, isomeri e CFT
advertisement

CH7. Intro to Coordination
Compounds
1
Inner-sphere vs outer-sphere
2
Nomenclature
1. Learn common ligand names (Table 7.1)
Ex:
:OH2
:O2
:CN
:Br
:NH3
aqua
oxo (oxido)
cyano (cyanido)
bromo (bromido)
ammine
Note that anionic ligands end in “o”
2. List ligands in alphabetical order
3. Metal name at end, add “ate” if it’s an anionic complex
some common names – ferrate, stannate, plumbate, cuprate
4. Add (and metal oxidation number in Roman numerals)
or add metal (and total complex charge in Arabic numerals)
3
Nomenclature
ex: [Cu(OH2)6]2+ is hexaaquacopper(II) or hexaaquacopper(2+)
[CuCl4]
is tetrachlorocuprate(III) or tetrachloridocuprate(III)
5. Add prefixes to indicate number of each ligand type
mono, di, tri, tetra, penta, hexa
or use bis, tris, tetrakis if less confusing due to ligand name
ex: [PtBr2{P(CH3)3}2 ] is dibromobis(trimethylphosphine)platinum(II)
~ C2v
~D2h
Stereoisomers
cis- and transplatin. The cis
isomer is an anticancer drug.
4
Cis-platin binding to DNA
5
Nomenclature
6. To write the formula:
[metal, then anionic ligands, then neutral ligands] net charge superscript
7.
Special ligands:
a. ambidentate
-SCN (thicyanato) vs NCS (isothiocyanato)
[Pt(SCN)4]2 D4h
[Cr(NCS)(NH3)5]2+
NO2 (nitrito)
vs
tetrathiocyanatoplatinate(II)
pentaammineisothiocyanatochromium(III)
ONO (isonitrito)
6
Nomenclature
b. bidentate – ligands bind to M at two sites
ex: H2NCH2CH2NH2 ethylenediamine (en)
[Cr(en)3]3+ tris(ethylenediamine)chromium(III)
View looking down C3 axis
D3 (-> no , no S axes, chiral)
enantiomers
7
Nomenclature
Another bidentate example is acetato
c. polydentate ligands – bind at multiple sites
ex: tetraazamacrocycles
porphine (a simple porphyrin)
the 4 N atoms are approximately square planar
8
Geometric Isomers
There have distinct physical and chemical properties
Oh coordination MX5Y
1 isomer
MX4Y2
2 isomers (cis or trans)
MX3Y3
2 isomers (fac = C3V or mer = C2V )
ex: [CoCl2(NH3)4]+ tetraamminedichlorocobalt(III)
cis – purple
trans – green
9
Optical Isomers
Enantiomers = non-superimposable mirror images of a
chiral molecule
enantiomers have identical physical properties (except in a
chiral environment, for example retention times on a chiral
column are not the same)
enantiomers rotate the plane of polarized light in opposite
directions (optical isomers)
10
Polymetallic complexes
(also called cage compounds)
no direct M-M bonding
ex:
MeOH (dry) / N2
S8 + NaSR + FeCl3
[Fe4S4(SR)4]n model for ferrodoxins
11
Cluster compounds
direct M-M bonding
ex:
[Re2Cl8]2 octachlorodirhenate(III)
D4h (eclipsed)
12
Crystal Field Theory
Oh complexes – put 6 e pairs around central metal in Oh geometry
this splits the 4 d-orbitals into 2 symmetry sets
t2g (xz, yz, xy)
and
eg (x2 – y2, z2)
0 can be determined from spectroscopic
data (see Table 8.3)
13
UV/Vis spectrum for Ti(OH2)63+
20,300 cm-1 (wavenumber units)
= 493 nm (wavelength units)
= 243 kJ/mol (energy units)
violet solution
14
Crystal Field Theory
0 depends on:
1. ligand (spectrochemical series)
0
I < Br < Cl < F < OH < NH3 < CN < CO
weak field
strong field
more complete list in text
2.
metal ion
0 greater for higher oxidation number – stronger, shorter M-L interaction
0 greater going down a group – more diffuse d-orbitals interact more
strongly with ligands
0
Mn2+ < Fe2+ < Fe3+ < Ru3+ < Pd4+ < Pt4+
small
large
15
Ligand Field Stabilization Energy
the LFSE = (0.4x 0.6y) 0
for electronic config t2gx egy
high spin case
# d electrons
0
1
2
3
4
e config
-
t2g1
t2g2
t2g3
LFSE (0)
0
0.4
0.8
# unpaired e
0
1
2
5
6
7
8
9
10
t2g3eg1 t2g3eg2 t4eg2
t5eg2
t2g6eg2 t2g6eg3 t2g6eg4
1.2
0.6
0
0.4
0.8
1.2
0.6
0
3
4
5
4
3
2
1
0
depends of relative values of 0 and pairing energy.
16
High spin vs low spin d4
t2g3eg1
t2g4
LSFE = 0.6 0
LFSE = 1.6 0 PE
high spin
low spin
(weak field)
(strong field)
[Cr(OH2)6]2+
[Cr(CN)6]4
17
Hhyd for first-row TM2+ ions
All are high spin complexes
H2O
M2+(g)
[M(OH2)6]2+ (aq)
H calc from Born Haber analyses
18
Magnetic Measurements
Magnetic moment () is the attractive force towards a magnetic field (H)
≈ [N(N + 2)]1/2 B
where N = number of unpaired electrons
N
/B
1
2
3
4
5
1.73
2.83
3.87
4.90
5.92
this is the paramagnetic contribution from unpaired e spin only, it ignores both spinorbit coupling and diamagnetic contributions
ex: [Mn(NCS)6]4 experimental /B = 6.06,
Mn(II) is d5 it must be a high spin complex
19
CN = 5
20
d-orbital splitting in a Td field
21
CFT for CN 4
For Td complexes
T << 0 due to fewer ligands and the geometry of field vs ligands
Δ
ex:
[CoCl4] 2
[Co(OH2)6]3+
3300 cm 1
20,700 cm 1
therefore Td complexes are nearly always high spin (pairing E more
important than LFSE)
Co(II) d7
ex:
LSFE = 1.2T
Fe3O4 magnetite Fe(II)Fe(III)2O4
oxide is a weak field ligand, so high spin case
Fe(II) is d6 (only in Oh sites); Fe(III) is d5 (1/2 in Oh sites, ½ in Td sites)
22
Tetragonal distortion of Oh
23
Square planar complexes
D4h is a common structure for d8 complexes (full z2, empty x2 – y2
orbitals)
Group 9: Rh(I), Ir(I)
Group 10: Pt(II), Pd(II)
Group 11: Au(III), for example AuCl4
Note: [Ni(CN)4]2 is D4h but [NiCl4]2 is Td
Ni(II) has a smaller than Pd, PT so Td is common
but we see D4h with strong field ligands
24
Jahn-Teller effect
Jahn-Teller effect: degenerate electronic ground states generate structural
disorder to decrease E
Ex: [Cu(OH2)6]2+
Cu(II) d9
We see a tetragonal distortion
But fluxional above 20K, so appears Oh by NMR in aqueous solution
25
Jahn-Teller effect
CuF2
26
Ligand Field Theory
CFT does not explain ligand
field strengths; MO theory
can
Start with SALCs that
are ligand combinations
shown to the right
27
MO for Oh TM complexes
SF6 - no metal d
valence orbitals
considered
28
p-bonding in Oh complexes
p-donor ligands
Decrease O
Example: Cl-
p-acceptor ligands
Increase O
Example: CO
29
Oh character table
30